Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication - PubMed (original) (raw)
Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication
Agnel Sfeir et al. Cell. 2009.
Abstract
Telomeres protect chromosome ends through the interaction of telomeric repeats with shelterin, a protein complex that represses DNA damage signaling and DNA repair reactions. The telomeric repeats are maintained by telomerase, which solves the end replication problem. We report that the TTAGGG repeat arrays of mammalian telomeres pose a challenge to the DNA replication machinery, giving rise to replication-dependent defects that resemble those of aphidicolin-induced common fragile sites. Gene deletion experiments showed that efficient duplication of telomeres requires the shelterin component TRF1. Without TRF1, telomeres activate the ATR kinase in S phase and show a fragile-site phenotype in metaphase. Single-molecule analysis of replicating telomeres showed that TRF1 promotes efficient replication of TTAGGG repeats and prevents fork stalling. Two helicases implicated in the removal of G4 DNA structures, BLM and RTEL1, were required to repress the fragile-telomere phenotype. These results identify a second telomere replication problem that is solved by the shelterin component TRF1.
Figures
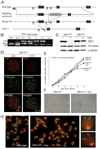
Figure 1. Conditional deletion of mouse TRF1
(A) Schematic of the mouse TRF1 locus on chromosome 1 (NCBI locus ID 21749), the targeting construct, and the altered alleles of TRF1. R: _EcoR_I, P: _Pvu_II, H: _Hind_III. F1. F2 and R: PCR primers. (B) TRF1 PCR on tail DNA from mice of the indicated genotypes using the F1 and F2 forward primers and the R reverse primer. PCR products: wild type, 100 bp; flox, 152 bp; null allele, 500 bp. (C) Immunoblot monitoring loss of TRF1 upon Cre-treatment of TRF1F/F MEFs. TRF1 (Ab1449) was detected 3 days after Cre treatment of TRF1F/F and TRF1 F/+ MEFs. -Cre: mock infection, γ-tubulin: loading control. (D) IF-FISH to monitor TRF1 at telomeres of TRF1F/F MEFs at day 3 post Cre. TRF1 IF, red; telomeric FITC PNA probe, green. (E) Graph representing proliferation of TRF1-deficient MEFs. (F) Phase-contrast microscopic images of primary TRF1F/F MEFs before and after Cre treatment. Cells were stained for SA-β-galactosidase at day 7 post Cre. (G) Metaphase spreads showing the fragile telomere phenotype in TRF1 null cells. Telomeres highlighted by FISH (green) and DNA was stained with DAPI (red) at day 4 post Cre.
Figure 2. Effects of TRF1, aphidicolin, and ATRsh
(A) Examples of the fragile telomeres in TRF1F/F MEF metaphases at day 4 post Cre, Telomeric DNA: FITC PNA probe (green); DNA: DAPI (blue). (B) Quantification of fragile telomeres induced by deletion of TRF1 with or without treatment with 0.2 µM aphidicolin or ATR shRNA. Bars represent mean values of three independent experiments with SDs. Asterisks: P < 0.01 based on a two-tailed Student's _t_-test. (C) Quantification chromosome breaks/gaps. Experimental conditions as in (B). (D) Quantification of long arms telomere associations in response to the indicated treatments. Experimental conditions as in (B). Sister telomere associations were only scored on long arms. Sister associations were significantly reduced (P < 0.05 based on a two-tailed Student's _t_-test) by treatment of TRF1 null cells with ATR shRNA but not by aphidicolin treatment or absence of DNA ligase IV (lig4−/−).
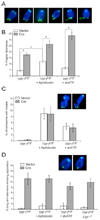
Figure 3. Deletion of TRF1 results in an S phase dependent ATR kinase signal
(A) ATR-dependent TIF formation upon deletion of TRF1 from cycling cells. Cells with the indicated genotypes were analyzed at day 4 post pWZL-Cre using FISH for telomeres (green), IF for 53BP1 (red), and DAPI DNA counterstain (blue). To circumvent the lethality associated with ATR deletion, f the TRF1F/F ATRF/F cells were arrested in G0 by contact inhibition and serum starvation, infected with Ad-Cre, released at day 3, and analyzed 1 day later. For the two right-hand panels, TRF1 F/− and TRF2 F/− cells were similarly arrested in G0 and infected with Ad-Cre, but analyzed at 4 day while in G0. Deletion of ATR, TRF1 and TRF2 was verified by immunoblotting (Suppl. Fig. 5). (B) Quantification of the TIF response as shown in panel (A). Bargraphs represent mean values of three independent experiments and SDs. Asterisks: P < 0.01 based on a two-tailed Student's _t_-test. (C) Immunoblot for Chk1 phosphorylation. Cells with the indicated genotypes were analyzed at day 6 post Cre. POT1a null MEFs and cells treated with UV (25 J/m2, 30 min recovery) serve as positive controls. (D) Immunoblot for Chk2 phosphorylation. Cells were treated as in (C). MEFs treated with IR (2 Gy, 1 hr recovery) serve as a positive control. (E) S phase dependent induction of TIFs. TRF1F/F cells were synchronized in G0 and infected with Ad-Cre and analyzed as in (A). For G1, cells were released into normal medium on day 3 post Cre and harvested 15 hrs post release. S/G2 cells were released into normal medium followed by an aphidicolin block and analyzed 7 hours after release from the G1/S block. Bargraphs represent mean values of three independent experiments and SDs. TRF1 was deleted in ∼50% of the cells (Suppl Fig. 5). FACS analysis of the G0, G1, and S/G2 cells is shown in Suppl Fig. 5.
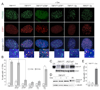
Figure 4. SMARD analysis of telomere replication in wild type cells
(A) Top: schematic depiction of the SMARD protocol to visualize the replication of single telomeric DNA molecules. See text for description. Bottom: Telomeric DNA molecules of variable lengths identified by telomeric FISH (TelC; blue) with incorporated IdU and CldU detected with fluorescent antibodies (red and green, respectively). The telomeric fragments are organized assuming that replication proceeds from a subtelomeric origin towards the chromosome end. (B) Two examples of replication fork progression towards the chromosome end. SMARD on ∼180-kb telomeric DNA _Swa_I fragments containing subtelomeric DNA of variable lengths. Procedure as in (A) except that the DNA was digested with _Swa_I and the DNA was resolved on a pulse-field gel (see genomic blot inset). Duration of the IdU and CldU pulses was 1 hour each. The pattern is consistent with replication forks progressing from a subtelomeric origin towards the chromosome end as depicted in the cartoon below each SMARD image. (C) Examples of three telomeric molecules with IdU/CldU incorporation patterns consistent with replication initiating in the TTAGGG sequence. Procedure as in (A).
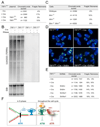
Figure 5. Deletion of TRF1 diminishes the replication efficiency of telomeric DNA
SMARD assay results from three independent experiments in which TRF1 was deleted from TRF1F/F MEFs. Cells were labeled with IdU and CldU as indicated on the right at day 4 after infection with H&R Cre retrovirus (+Cre) or vector control (− Cre). In the upper panel (experiments 1 and 2), DNA was digested with frequently cutting enzymes and telomeric restriction fragments >25 kb were isolated (schematic on the left). Telomeric DNA molecules were identified by FISH and the % of molecules containing IdU and/or CldU was determined. In the middle panel, telomeric _Mbo_I/_Alu_I fragments in the 130–180 kb range (see genomic blot inset) were isolated from a CHEF gel and the fraction of telomeric molecules that contained IdU and/or CldU was determined as above. SMARD assay was done in one experiment in which TRF2 was deleted from TRF2F/F _DNA-Lig4_−/− cells and the fraction IdU and/or CldU labeled telomeric molecules (130–180 kb range) was analyzed. In the lower panel, the DNA preparation of TRF1F/F MEFs used in the middle panel was digested with _Swa_I and a 180 kb restriction fragment from the Igh locus was isolated. DNA probes from that locus (see map below) were used to identify the Igh fragments on stretched DNA and the ratio of labeled vs. unlabeled fragments was determined.
Figure 6. Evidence for fork stalling at telomeres in cells lacking TRF1
(A) Evidence for replication fork stalling at the subtelomeric/telomeric boundary. Shown are four _Swa_I DNA fragments containing telomeric repeats and subtelomeric DNA from cells lacking TRF1. For procedure see Fig. 4. Molecules shown represent incorporation patterns of IdU and CldU consistent with replication of the subtelomeric DNA (lacking TelC FISH signal) and fork stalling at the boundary of subtelomeric and telomeric DNA as shown in the schematic. (B) Evidence for replication fork stalling after initiation of DNA replication within telomeric DNA in TRF1 deficient cells. DNA was cut with frequently cutting restriction enzymes and molecules >25 kb were isolated. Labeling was performed as in Fig. 5, top panel. The patterns of incorporation of IdU and CldU are consistent with initiation of replication within the telomeric DNA near the end of a IdU pulse followed by fork progression in only one direction during the CldU labeling period.
Figure 7. The mechanism by which TRF1 represses telomere fragility
(A) Frequency of fragile telomeres in Cre-treated TRF1F/F MEFs complemented with TRF1ΔAc or TRF1ΔMyb (see Suppl. Fig. 6A for metaphase spreads and TRF1 immunoblots). (B) TERRA detected by Northern blotting of RNA from cells with the indicated genotype at day 4 post Cre. Bottom: Ethidium bromide (EtBr) staining of rRNAs serves as loading control. (C) Fragile telomere incidence in cells lacking Blm and/or Wrn. (D) Representative metaphase spreads from TRF1F/F MEFs (+ or − Cre treatment) infected with Blm and Rtel1 shRNAs as indicated. (E) Quantification of fragile telomeres in TRF1F/F MEFs (+ or − Cre treatment) infected with Blm and Rtel1 shRNAs as indicated. See Suppl. Fig. 6 for validation of the shRNAs. (F) Schematic summarizing the proposed function of TRF1 in the context of shelterin. While TRF2 and POT1 repress the DNA damage response throughout the cell cycle, TRF1 is proposed to act in S phase to facilitate replication fork progression through the telomeric DNA. TRF1 is proposed to prevent formation of a fork barrier (most likely G4 DNA) in part by acting with BLM and RTEL1.
Similar articles
- TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling.
Zimmermann M, Kibe T, Kabir S, de Lange T. Zimmermann M, et al. Genes Dev. 2014 Nov 15;28(22):2477-91. doi: 10.1101/gad.251611.114. Epub 2014 Oct 24. Genes Dev. 2014. PMID: 25344324 Free PMC article. - A TRF1-controlled common fragile site containing interstitial telomeric sequences.
Bosco N, de Lange T. Bosco N, et al. Chromosoma. 2012 Oct;121(5):465-74. doi: 10.1007/s00412-012-0377-6. Epub 2012 Jul 13. Chromosoma. 2012. PMID: 22790221 Free PMC article. - TopoIIα prevents telomere fragility and formation of ultra thin DNA bridges during mitosis through TRF1-dependent binding to telomeres.
d'Alcontres MS, Palacios JA, Mejias D, Blasco MA. d'Alcontres MS, et al. Cell Cycle. 2014;13(9):1463-81. doi: 10.4161/cc.28419. Epub 2014 Mar 10. Cell Cycle. 2014. PMID: 24626180 Free PMC article. - TFIIH moonlighting at telomeres.
Glousker G, Lingner J. Glousker G, et al. Genes Dev. 2022 Sep 1;36(17-18):951-953. doi: 10.1101/gad.350140.122. Genes Dev. 2022. PMID: 36347559 Free PMC article. Review. - Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance.
Walker JR, Zhu XD. Walker JR, et al. Mech Ageing Dev. 2012 Jun;133(6):421-34. doi: 10.1016/j.mad.2012.05.002. Epub 2012 May 23. Mech Ageing Dev. 2012. PMID: 22634377 Review.
Cited by
- ATR cooperates with CTC1 and STN1 to maintain telomeres and genome integrity in Arabidopsis.
Boltz KA, Leehy K, Song X, Nelson AD, Shippen DE. Boltz KA, et al. Mol Biol Cell. 2012 Apr;23(8):1558-68. doi: 10.1091/mbc.E11-12-1002. Epub 2012 Feb 22. Mol Biol Cell. 2012. PMID: 22357613 Free PMC article. - Generation of a mouse model for studying the role of upregulated RTEL1 activity in tumorigenesis.
Wu X, Sandhu S, Nabi Z, Ding H. Wu X, et al. Transgenic Res. 2012 Oct;21(5):1109-15. doi: 10.1007/s11248-011-9586-7. Epub 2012 Jan 12. Transgenic Res. 2012. PMID: 22238064 Free PMC article. - Cohesin Mutations in Cancer.
De Koninck M, Losada A. De Koninck M, et al. Cold Spring Harb Perspect Med. 2016 Dec 1;6(12):a026476. doi: 10.1101/cshperspect.a026476. Cold Spring Harb Perspect Med. 2016. PMID: 27742736 Free PMC article. Review. - Replication fork stalling in late S-phase elicits nascent strand degradation by DNA mismatch repair.
Colicino-Murbach E, Hathaway C, Dungrawala H. Colicino-Murbach E, et al. Nucleic Acids Res. 2024 Oct 14;52(18):10999-11013. doi: 10.1093/nar/gkae721. Nucleic Acids Res. 2024. PMID: 39180395 Free PMC article. - Budding yeast Rap1, but not telomeric DNA, is inhibitory for multiple stages of DNA replication in vitro.
Douglas ME, Diffley JFX. Douglas ME, et al. Nucleic Acids Res. 2021 Jun 4;49(10):5671-5683. doi: 10.1093/nar/gkab416. Nucleic Acids Res. 2021. PMID: 34048583 Free PMC article.
References
- Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA076027-11/CA/NCI NIH HHS/United States
- R37 GM049046/GM/NIGMS NIH HHS/United States
- R01 AG016642/AG/NIA NIH HHS/United States
- R37 GM049046-17/GM/NIGMS NIH HHS/United States
- R01 CA076027/CA/NCI NIH HHS/United States
- R01 AG016642-10/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous