A dynamic network of transcription in LPS-treated human subjects - PubMed (original) (raw)
A dynamic network of transcription in LPS-treated human subjects
Junhee Seok et al. BMC Syst Biol. 2009.
Abstract
Background: Understanding the transcriptional regulatory networks that map out the coordinated dynamic responses of signaling proteins, transcription factors and target genes over time would represent a significant advance in the application of genome wide expression analysis. The primary challenge is monitoring transcription factor activities over time, which is not yet available at the large scale. Instead, there have been several developments to estimate activities computationally. For example, Network Component Analysis (NCA) is an approach that can predict transcription factor activities over time as well as the relative regulatory influence of factors on each target gene.
Results: In this study, we analyzed a gene expression data set in blood leukocytes from human subjects administered with lipopolysaccharide (LPS), a prototypical inflammatory challenge, in the context of a reconstructed regulatory network including 10 transcription factors, 99 target genes and 149 regulatory interactions. We found that the computationally estimated activities were well correlated to their coordinated action. Furthermore, we found that clustering the genes in the context of regulatory influences greatly facilitated interpretation of the expression data, as clusters of gene expression corresponded to the activity of specific factors or more interestingly, factor combinations which suggest coordinated regulation of gene expression. The resulting clusters were therefore more biologically meaningful, and also led to identification of additional genes under the same regulation.
Conclusion: Using NCA, we were able to build a network that accounted for between 8-11% genes in the known transcriptional response to LPS in humans. The dynamic network illustrated changes of transcription factor activities and gene expressions as well as interactions of signaling proteins, transcription factors and target genes.
Figures
Figure 1
Schematic of the approach. (A) Flowchart describing the steps to reconstruct our initial transcriptional regulatory network. (B) A set of gene expression profiles (matrix E) and about a proposed structure for the underlying transcriptional regulatory network (matrix S(0)) are used as inputs for Network Component Analysis (NCA). NCA uses an algorithm that first calculates the expected transcription factor activities (matrix A), and then recalculates S based on the new values of A, until both matrices converge. The outputs of this procedure are A* and S*, final values of A and S, which provide information about transcription factor activity as well as regulatory structure, respectively.
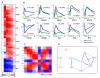
Figure 2
Transcription factor activities calculated using NCA. (A) Predicted activities of the ten transcription factors used in this study. For each transcription factor, rows represent progression in time and columns correspond to the four human subjects. Activities of each row are normalized to the zero time point. (B) Transcription factor activities (blue) compared to gene expression (green), with Pearson correlation coefficients noted. Both activity and expression at each time point are averages normalized to the time = 0 values, and the activity is further scaled for direct comparison with the expression values. (C) Correlation matrix between transcription factor activities. Red represents positive correlation, and blue represents negative correlation. (D) Inferred combinatorial regulation pairs of transcription factors. A blue solid line indicates that the pair was supported by protein-protein interaction knowledge of BIND and high correlation of their activities (>0.75). A black solid line indicates that the pair was only supported by high correlation, and a blue dotted line indicates that the pair was only supported by the interaction database.
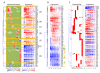
Figure 3
Hierarchical clustering in the context of a defined regulatory network. (A) The adjusted strength matrix was used for clustering, after which the gene expression matrix was appended. Seven major clusters which have more than five associated genes are highlighted. In the adjusted strength matrix heatmap, green color indicates that there is no prior regulatory connection in our model while white color indicates a weak regulatory influence. (B) Clustering with gene expression only. Genes in the Cluster F(regulated by STAT6) were noted with green dots, and genes in the Cluster G(regulated by MYC) were noted with orange dots. (C) Clustering with the binary regulatory relations (initial connectivity matrix) assuming all regulatory strengths are equal.
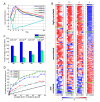
Figure 4
Identification of new target genes for major clusters. (A) The average expression profiles of the four clusters with > 10 members. (B) Expressions of extended regulatory genes sorted by correlation coefficients(c) with the average expression profile of a cluster. Each extended gene group was divided into highly correlated (c > 0.5), un-correlated (t0.5 <_c_ < 0.5) and anti-correlated (_c_ < t0.5) groups. The average gene expression of each cluster is shown as a row at the top of each column. (C) Ratio of highly correlated genes (_c_ > 0.5) in the sets of extended regulatory genes and 1,000 randomly chosen genes. Error bars were calculated as the standard deviation of a population derived from 100 repeated tests. P-values measured by the Fisher's exact test are noted above each column set. (D) Fraction of acceptable new predicted cluster genes from both the extended and "no evidence" gene sets. Significantly expressed genes (p < 0.01) in both sets were plotted against each other using a range of Pearson's coefficient cutoff values for Clusters A, B, D, and G. The dashed line indicates where the fraction of acceptable genes is equal from both the extended and "no evidence" sets
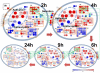
Figure 5
A dynamic network of transcription. At time zero, LPS is injected, giving rise to transcription factor activation, which then leads to induction or repression of gene expression, production and secretion of cytokines, and initiation of secondary signals. Target genes which correspond to secreted proteins (e.g., IL10, IL1A and IL1B) are noted with green circles, and transcription factors that are regulated by other factors, such as STAT1 and MYC, are noted with cyan circles. The seven major clusters marked in Figure 3A are grouped with orange boxes. Black lines denote activation of a transcription factor by an extracellular signal, red and blue lines show the influence of a transcription factor on a target gene, and green dotted lines indicate secretion of a gene product.
Similar articles
- A transcriptional dynamic network during Arabidopsis thaliana pollen development.
Wang J, Qiu X, Li Y, Deng Y, Shi T. Wang J, et al. BMC Syst Biol. 2011;5 Suppl 3(Suppl 3):S8. doi: 10.1186/1752-0509-5-S3-S8. Epub 2011 Dec 23. BMC Syst Biol. 2011. PMID: 22784627 Free PMC article. - Inferring a transcriptional regulatory network of the cytokinesis-related genes by network component analysis.
Chen SF, Juang YL, Chou WK, Lai JM, Huang CY, Kao CY, Wang FS. Chen SF, et al. BMC Syst Biol. 2009 Nov 27;3:110. doi: 10.1186/1752-0509-3-110. BMC Syst Biol. 2009. PMID: 19943917 Free PMC article. - Dynamic transcriptional control of macrophage miRNA signature via inflammation responsive enhancers revealed using a combination of next generation sequencing-based approaches.
Czimmerer Z, Horvath A, Daniel B, Nagy G, Cuaranta-Monroy I, Kiss M, Kolostyak Z, Poliska S, Steiner L, Giannakis N, Varga T, Nagy L. Czimmerer Z, et al. Biochim Biophys Acta Gene Regul Mech. 2018 Jan;1861(1):14-28. doi: 10.1016/j.bbagrm.2017.11.003. Epub 2017 Nov 11. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29133016 - An Overview of NCA-Based Algorithms for Transcriptional Regulatory Network Inference.
Wang X, Alshawaqfeh M, Dang X, Wajid B, Noor A, Qaraqe M, Serpedin E. Wang X, et al. Microarrays (Basel). 2015 Nov 16;4(4):596-617. doi: 10.3390/microarrays4040596. Microarrays (Basel). 2015. PMID: 27600242 Free PMC article. Review. - Time-Based Systems Biology Approaches to Capture and Model Dynamic Gene Regulatory Networks.
Alvarez JM, Brooks MD, Swift J, Coruzzi GM. Alvarez JM, et al. Annu Rev Plant Biol. 2021 Jun 17;72:105-131. doi: 10.1146/annurev-arplant-081320-090914. Epub 2021 Mar 5. Annu Rev Plant Biol. 2021. PMID: 33667112 Free PMC article. Review.
Cited by
- A systems biology approach to the analysis of subset-specific responses to lipopolysaccharide in dendritic cells.
Hancock DG, Shklovskaya E, Guy TV, Falsafi R, Fjell CD, Ritchie W, Hancock RE, Fazekas de St Groth B. Hancock DG, et al. PLoS One. 2014 Jun 20;9(6):e100613. doi: 10.1371/journal.pone.0100613. eCollection 2014. PLoS One. 2014. PMID: 24949855 Free PMC article. - Recent progress using systems biology approaches to better understand molecular mechanisms of immunity.
Gottschalk RA, Martins AJ, Sjoelund V, Angermann BR, Lin B, Germain RN. Gottschalk RA, et al. Semin Immunol. 2013 Oct 31;25(3):201-8. doi: 10.1016/j.smim.2012.11.002. Epub 2012 Dec 11. Semin Immunol. 2013. PMID: 23238271 Free PMC article. Review. - Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells.
St Laurent G, Shtokalo D, Tackett MR, Yang Z, Eremina T, Wahlestedt C, Urcuqui-Inchima S, Seilheimer B, McCaffrey TA, Kapranov P. St Laurent G, et al. BMC Genomics. 2012 Sep 24;13:504. doi: 10.1186/1471-2164-13-504. BMC Genomics. 2012. PMID: 23006825 Free PMC article. - Spatiotemporal positioning of multipotent modules in diverse biological networks.
Chen Y, Wang Z, Wang Y. Chen Y, et al. Cell Mol Life Sci. 2014 Jul;71(14):2605-24. doi: 10.1007/s00018-013-1547-2. Epub 2014 Jan 11. Cell Mol Life Sci. 2014. PMID: 24413666 Free PMC article. Review. - Dynamic regulatory network reconstruction for Alzheimer's disease based on matrix decomposition techniques.
Kong W, Mou X, Zhi X, Zhang X, Yang Y. Kong W, et al. Comput Math Methods Med. 2014;2014:891761. doi: 10.1155/2014/891761. Epub 2014 Jun 15. Comput Math Methods Med. 2014. PMID: 25024739 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- U54 GM062119/GM/NIGMS NIH HHS/United States
- K99 CA125994/CA/NCI NIH HHS/United States
- U54-GM62119/GM/NIGMS NIH HHS/United States
- R00 CA125994/CA/NCI NIH HHS/United States
- CA125994-01A1/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources