Focal distribution of hepatitis C virus RNA in infected livers - PubMed (original) (raw)
Focal distribution of hepatitis C virus RNA in infected livers
J David Stiffler et al. PLoS One. 2009.
Abstract
Background: Hepatitis C virus (HCV) is a plus-strand RNA virus that replicates by amplification of genomic RNA from minus strands leading to accumulation of almost one thousand copies per cell under in vitro cell culture conditions. In contrast, HCV RNA copy numbers in livers of infected patients appear to be much lower, estimated at a few copies per cell.
Methodology/principal findings: To gain insights into mechanisms that control HCV replication in vivo, we analyzed HCV RNA levels as well as expression of interferon beta (IFNbeta) and several interferon stimulated genes (ISGs) from whole liver sections and micro-dissected subpopulations of hepatocytes in biopsy samples from 21 HCV-infected patients. The results showed that intrahepatic HCV RNA levels range form less than one copy per hepatocyte to a maximum of about eight. A correlation existed between viral RNA levels and IFNbeta expression, but not between viral RNA and ISG levels. Also, IFNbeta expression did not correlate with ISGs levels. Replication of HCV RNA occurred in focal areas in the liver in the presence of a general induction of ISGs.
Conclusion/significance: The low average levels of HCV RNA in biopsy samples can be explained by focal distribution of infected hepatocytes. HCV replication directly induces IFNbeta, which then activates ISGs. The apparent lack of a correlation between levels of IFNbeta and ISG expression indicates that control of the innate immune response during HCV infections depends on multiple factors.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
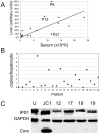
Figure 1. Correlation between HCV RNA in serum and liver.
(A) Values for viral RNA in serum (x-axis) were plotted against HCV RNA levels (y-axis) in frozen sections from needle biopsies determined by quantitative real-time PCR (qRT-PCR). The values were normalized with albumin and expressed on a scale from 0 to 100. (B) The plot shows the estimated copy number of HCV RNA per hepatocyte for each of the 22 patients listed in Table 1. C) The figure shows a western blot analysis with protein extracts from normal, uninfected Huh7 cells (U), HCV JC1 infected Huh7 cells (JC1) and from liver tissues of patients 12, 17,18 and 19 (see Table 1). The blot was incubated with antibodies specific for the indicated proteins. Core, HCV core protein.
Figure 2. Correlation between HCV RNA and activation of IFNβ and ISGs.
The Ct values for HCV, IFNβ, Mx1 and IFIT1 obtained from real-time qRT-PCR were plotted in different combinations as indicated. Ctn represents Ct values that were normalized with the Ct for albumin to account for the difference in cell number on each frozen section used for RNA isolation. Note, that Ct values and concentration of RNA are inversely related and that a difference of 1 in the Ct value corresponds to a two-fold difference in concentration.
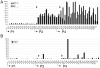
Figure 3. Focal accumulation of HCV RNA.
A) The bar graph shows the levels of Mx1 and IFIT1 expression in groups of about 100 cells that were isolated by LCM from frozen section of patients P1, P2 and P4. Ct values obtained for the indicated genes were normalized with Ct for18S RNA. Expression levels are expressed on a scale from 0 to 100. B) Same as A except that the values for HCV RNA were displayed. The samples were sorted in order of the patients (P1, P2, P4) as indicated in the figure.
Similar articles
- Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver.
Wieland S, Makowska Z, Campana B, Calabrese D, Dill MT, Chung J, Chisari FV, Heim MH. Wieland S, et al. Hepatology. 2014 Jun;59(6):2121-30. doi: 10.1002/hep.26770. Epub 2014 Apr 25. Hepatology. 2014. PMID: 24122862 Free PMC article. - Use of laser capture microdissection to map hepatitis C virus-positive hepatocytes in human liver.
Kandathil AJ, Graw F, Quinn J, Hwang HS, Torbenson M, Perelson AS, Ray SC, Thomas DL, Ribeiro RM, Balagopal A. Kandathil AJ, et al. Gastroenterology. 2013 Dec;145(6):1404-13.e1-10. doi: 10.1053/j.gastro.2013.08.034. Epub 2013 Aug 22. Gastroenterology. 2013. PMID: 23973767 Free PMC article. - High resolution sequencing of hepatitis C virus reveals limited intra-hepatic compartmentalization in end-stage liver disease.
Hedegaard DL, Tully DC, Rowe IA, Reynolds GM, Bean DJ, Hu K, Davis C, Wilhelm A, Ogilvie CB, Power KA, Tarr AW, Kelly D, Allen TM, Balfe P, McKeating JA. Hedegaard DL, et al. J Hepatol. 2017 Jan;66(1):28-38. doi: 10.1016/j.jhep.2016.07.048. Epub 2016 Aug 13. J Hepatol. 2017. PMID: 27531641 Free PMC article. - Innate and adaptive immune responses in HCV infections.
Heim MH, Thimme R. Heim MH, et al. J Hepatol. 2014 Nov;61(1 Suppl):S14-25. doi: 10.1016/j.jhep.2014.06.035. Epub 2014 Nov 3. J Hepatol. 2014. PMID: 25443342 Review. - [HCV-RNA in liver of chronic hepatitis C--plus strand RNA and minus strand RNA].
Endo H, Yamada G. Endo H, et al. Nihon Rinsho. 1995 Sep;53 Suppl(Pt 1):623-8. Nihon Rinsho. 1995. PMID: 7563844 Review. Japanese. No abstract available.
Cited by
- Hepatocytes and the art of killing Plasmodium softly.
Marques-da-Silva C, Schmidt-Silva C, Kurup SP. Marques-da-Silva C, et al. Trends Parasitol. 2024 Jun;40(6):466-476. doi: 10.1016/j.pt.2024.04.004. Epub 2024 May 6. Trends Parasitol. 2024. PMID: 38714463 Review. - MicroRNA-122 Regulation of HCV Infections: Insights from Studies of miR-122-Independent Replication.
Panigrahi M, Palmer MA, Wilson JA. Panigrahi M, et al. Pathogens. 2022 Sep 2;11(9):1005. doi: 10.3390/pathogens11091005. Pathogens. 2022. PMID: 36145436 Free PMC article. Review. - MicroRNA 122 Affects both the Initiation and the Maintenance of Hepatitis C Virus Infections.
Panigrahi M, Thibault PA, Wilson JA. Panigrahi M, et al. J Virol. 2022 Feb 23;96(4):e0190321. doi: 10.1128/JVI.01903-21. Epub 2021 Dec 15. J Virol. 2022. PMID: 34908444 Free PMC article. - HCV Spread Kinetics Reveal Varying Contributions of Transmission Modes to Infection Dynamics.
Durso-Cain K, Kumberger P, Schälte Y, Fink T, Dahari H, Hasenauer J, Uprichard SL, Graw F. Durso-Cain K, et al. Viruses. 2021 Jul 6;13(7):1308. doi: 10.3390/v13071308. Viruses. 2021. PMID: 34372514 Free PMC article. - HIV influences clustering and intracellular replication of hepatitis C virus.
Goyal A, Perelson AS, Kandathil AJ, Quinn J, Balagopal A, Ribeiro RM. Goyal A, et al. J Viral Hepat. 2021 Feb;28(2):334-344. doi: 10.1111/jvh.13429. Epub 2020 Nov 24. J Viral Hepat. 2021. PMID: 33128322 Free PMC article.
References
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. - PubMed
- Berenguer M, Prieto M, San Juan F, Rayon JM, Martinez F, et al. Contribution of donor age to the recent decrease in patient survival among HCV-infected liver transplant recipients. Hepatology. 2002;36:202–210. - PubMed
- Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1101–1152.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources