Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA - PubMed (original) (raw)
Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA
Ling-Ling Chen et al. Mol Cell. 2009.
Abstract
In many cells, mRNAs containing inverted repeats (Alu repeats in humans) in their 3' untranslated regions (3'UTRs) are inefficiently exported to the cytoplasm. Nuclear retention correlates with adenosine-to-inosine editing and is in paraspeckle-associated complexes containing the proteins p54(nrb), PSF, and PSP1 alpha. We report that robust editing activity in human embryonic stem cells (hESCs) does not lead to nuclear retention. p54(nrb), PSF, and PSP1 alpha are all expressed in hESCs, but paraspeckles are absent and only appear upon differentiation. Paraspeckle assembly and function depend on expression of a long nuclear-retained noncoding RNA, NEAT1. This RNA is not detectable in hESCs but is induced upon differentiation. Knockdown of NEAT1 in HeLa cells results both in loss of paraspeckles and in enhanced nucleocytoplasmic export of mRNAs containing inverted Alu repeats. Taken together, these results assign a biological function to a large noncoding nuclear RNA in the regulation of mRNA export.
Figures
Figure 1
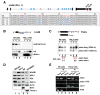
Altered nuclear retention in human ES cells. A. ADAR1 activity in hESCs. The upper panel shows the distribution of Alu elements in the Lin28 transcript. The lower panel shows the sequence of several cDNA clones of Lin28 showing A-to-G changes diagnostic of A-to-I editing. Red arrows indicate the primers used for amplification. B. Lin28 mRNA escapes nuclear retention in H9 cells. Nuclear and cytoplasmic RNAs were isolated from H9 cells and HeLa cells and resolved on a denaturing agarose gel. The probe for the northern blot (red bar) is located upstream of the IR_Alu_s from the 3′-UTR of Lin28. tRNAlys and U6 were used as makers for nuclear and cytoplasmic RNA fractionation. Equivalent fractions of total (T), cytoplasmic (C) and nuclear (N) RNAs were loaded. C. Paics mRNA escapes nuclear retention in H9 cells, but is mostly retained in the nucleus in HeLa cells. The same samples from panel B were used. Two Paics isoforms are shown. The probe for the northern blot (red bar) is located upstream of the IR_Alu_s from the 3′-UTR of Paics. Bands were quantitated using ImageJ software. D. ADAR1 and paraspeckle-related proteins are expressed in H9 cells. Total proteins were collected from HEK293 cells (Lane 1), HeLa cells (Lane 2), H9 cells (Lane 3) and H9 cells differentiated into trophoblasts using BMP4 (Lane 4). Actin was used as loading control, Sox2 was used as a marker for human ES cells and Troma-1 was used as a marker for trophoblasts. E. mRNAs containing IR_Alu_s are associated with p54nrbin both H9 cells and HeLa cells. Immunoprecipitations (IP) were performed from H9 cells and HeLa cells using anti-p54nrb antibody or mock antibody.
Figure 2
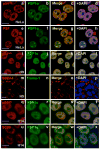
Human ES cells lack paraspeckles. HeLa cells were stained with anti-p54nrb and anti-PSP1α antibodies (a–d) or anti-PSFand anti-PSP1α antibodies (e–h). DAPI was used to indicate DNA. Note that p54nrb, PSF, and PSP1α colocalize with each other and indicate paraspeckles. p54nrb and PSP1α colocalize throughout the nucleoplasm in H9 cells, but show no apparent nuclear paraspeckles (i–l). The same batch of H9 cells was also stained with anti-SSEA4 (stem cell marker) and anti-Troma-1 (trophoblast differentiation marker) (m–p). p54nrb and PSP1α also colocalize throughout the nucleoplasm in H14 cells (q–t), but show no apparent nuclear paraspeckles. H14 cells were also stained with anti-SC35 and anti-PSP1α (u–x). The white scale bar in all images denotes 10 μm.
Figure 3
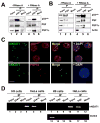
A. RNA-independent interaction of p54nrb with PSF and PSP1α in H9 cells. Total extracts of H9 cells with or without RNaseA treatment were immunoprecipitated with p54nrb antibody or mock antibody, and then immunoblotted with anti-p54nrb anti-PSF and anti-PSP1α antibodies. B. The same immunoprecipitation experiments were performed with anti-PSF antibody in H9 cells. Anti-actin was used as a negative control. C. RNA in situ hybridization was performed with green-dUTP-labeled antisense hNEAT1 probe in HeLa cells, and representative images are shown. p54nrband PSP1α are in red. hNEAT1 colocalizes with p54nrb (upper panel) and PSP1α (lower panel). D. hNEAT1 associates with p54nrband PSF in HeLa cells. IP with anti-p54nrb, anti-PSF, or mock antibody was carried out in extracts from H9 cells or HeLa cells. RT-PCR of hNEAT1 from the IP showed amplification in HeLa cells by anti-p54nrb IP and anti-PSF IP, but not by IP with mock antibody. Oct3/4 is a marker of H9 cells.
Figure 4
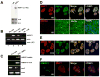
hNEAT1 and paraspeckles appear upon hESC differentiation. A. Northern blot showing absence of hNEAT1 RNA in H9 cells. B. RT-PCR showing absence of hNEAT1 RNA in H9 cells (lane 1, H9 cells cultured in mTeSR; lane 4, H9 cells cultured in CM medium), but its presence in differentiated cells (Lane 3). C. RT-PCR showing hNEAT1 absence in undifferentiated H14 cells, but expression in differentiated H14 cells. D. H9 cells cultured in CM medium were induced with BMP4 for 5 days, and then fixed for immunostaining. Day 5 differentiated H9 cells show the trophoblast marker, Troma-1 and the loss of stem cell marker, SSEA4 (lower panel). p54nrb and PSP1α colocalize with each other and form multiple large accumulations (white arrows), indicating paraspeckles. E. Day 4 differentiated H14 cells show the p54nrb and PSP1α accumulations (white arrows) in the nuclei, indicating paraspeckles. F. hNEAT1 (in situ hybridization, green) is expressed in trophoblasts derived from H9 cells and colocalizes with p54nrb.
Figure 5
Role of hNEAT1 in paraspeckle function. A. hNEAT1 RNA in situ hybridization indicates efficient knockdown of hNEAT1 levels after 48 hr treatment with pooled antisense (AS) oligodeoxynucleotides. B. hNEAT1 RNA correlates with paraspeckle formation. A typical staining pattern of hNEAT1 and p54nrb, (upper panel) and of hNEAT1 and PSP1α (lower panel) in hNEAT1 knockdown HeLa cells. Circled nuclei show that fewer paraspeckles are apparent in cells in which hNEAT1 has been knocked down. C. IR_Alu_s-containing mRNAs escape nuclear retention in hNEAT1 knockdown HeLa cells. Nuclear and cytoplasmic RNAs were isolated from untreated and hNEAT1 AS-treated HeLa cells and analyzed by RT-PCR. PCR primers are located in the downstream of IR_Alu_s in the 3′-UTRs to detect only full-length mRNAs (Fig. S3 and Fig. 5E, DP). In untreated HeLa cells, the majority of full-length Nup43, Paics, and Pccb mRNAs are retained in the nucleus (left panel), while significantly more is exported to the cytoplasm in hNEAT1 knockdown cells (right panel). Bands were quantitated using ImageJ software. In each case, omission of RT led to no signal (data not shown). T, total RNA; C, cytoplasmic RNA; N, nuclear RNA. D. Northern blotting. Nuclear and cytoplasmic RNAs were isolated from untreated and hNEAT1-AS treated HeLa cells and resolved on a denaturing agarose gel. In untreated HeLa cells, the majority of Paics mRNA is retained in the nucleus (left panel), while more Paics is exported to the cytoplasm in the hNEAT1 knockdown cells (right panel). The membrane was stripped and re-probed for actin mRNA, which shows no change in its cellular distribution after AS treatment. E. IR_Alu_s-containing mRNAs escape nuclear retention in HeLa cells after anti-hNEAT1 siRNA treatment. The upper panel shows two isoforms of Nup43, Paics, and Pccb: the long isoform has one pair of IR_Alu_s in its 3′-UTR, while the short one lacks IR_Alu_s. The red line indicates the location of the upstream probe (UP) which recognizes both isoforms, and the dark blue line indicates the location of the downstream probe (DP) which only recognizes the long IR_Alu_s-containing isoform. F. Controls. The same mRNA samples from Panel E were used. Smc1a, flnb, hyou1, and son are mRNAs that do not contain IR_Alu_s in their 3′-UTRs, but are normally enriched in the nucleus. These mRNAs show no change in their N/C distribution after hNEAT1 knockdown.
Figure 6
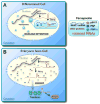
A. A model for nuclear retention of promiscuously edited RNAs in differentiated cells. Nuclear noncoding hNEAT1 RNA plays a critical role in paraspeckle formation and function. B. Altered gene regulation in human ES cells. In these cells, edited mRNAs are exported to the cytoplasm because of a defect in paraspeckle formation and function.
Comment in
- A NEAT way of regulating nuclear export of mRNAs.
Scadden D. Scadden D. Mol Cell. 2009 Aug 28;35(4):395-6. doi: 10.1016/j.molcel.2009.08.005. Mol Cell. 2009. PMID: 19716782
Similar articles
- A NEAT way of regulating nuclear export of mRNAs.
Scadden D. Scadden D. Mol Cell. 2009 Aug 28;35(4):395-6. doi: 10.1016/j.molcel.2009.08.005. Mol Cell. 2009. PMID: 19716782 - Protein arginine methyltransferase CARM1 attenuates the paraspeckle-mediated nuclear retention of mRNAs containing IRAlus.
Hu SB, Xiang JF, Li X, Xu Y, Xue W, Huang M, Wong CC, Sagum CA, Bedford MT, Yang L, Cheng D, Chen LL. Hu SB, et al. Genes Dev. 2015 Mar 15;29(6):630-45. doi: 10.1101/gad.257048.114. Genes Dev. 2015. PMID: 25792598 Free PMC article. - Alu element-mediated gene silencing.
Chen LL, DeCerbo JN, Carmichael GG. Chen LL, et al. EMBO J. 2008 Jun 18;27(12):1694-705. doi: 10.1038/emboj.2008.94. Epub 2008 May 22. EMBO J. 2008. PMID: 18497743 Free PMC article. - Paraspeckles: nuclear bodies built on long noncoding RNA.
Bond CS, Fox AH. Bond CS, et al. J Cell Biol. 2009 Sep 7;186(5):637-44. doi: 10.1083/jcb.200906113. Epub 2009 Aug 31. J Cell Biol. 2009. PMID: 19720872 Free PMC article. Review. - Paraspeckles as rhythmic nuclear mRNA anchorages responsible for circadian gene expression.
Torres M, Becquet D, Blanchard MP, Guillen S, Boyer B, Moreno M, Franc JL, François-Bellan AM. Torres M, et al. Nucleus. 2017 May 4;8(3):249-254. doi: 10.1080/19491034.2016.1277304. Epub 2017 Jan 6. Nucleus. 2017. PMID: 28060565 Free PMC article. Review.
Cited by
- Alternative cleavage and polyadenylation: the long and short of it.
Tian B, Manley JL. Tian B, et al. Trends Biochem Sci. 2013 Jun;38(6):312-20. doi: 10.1016/j.tibs.2013.03.005. Epub 2013 Apr 27. Trends Biochem Sci. 2013. PMID: 23632313 Free PMC article. Review. - Malat1 is not an essential component of nuclear speckles in mice.
Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Nakagawa S, et al. RNA. 2012 Aug;18(8):1487-99. doi: 10.1261/rna.033217.112. Epub 2012 Jun 20. RNA. 2012. PMID: 22718948 Free PMC article. - A toolkit for the identification of NEAT1_2/paraspeckle modulators.
An H, Elvers KT, Gillespie JA, Jones K, Atack JR, Grubisha O, Shelkovnikova TA. An H, et al. Nucleic Acids Res. 2022 Nov 11;50(20):e119. doi: 10.1093/nar/gkac771. Nucleic Acids Res. 2022. PMID: 36099417 Free PMC article. - Distinct roles of DBHS family members in the circadian transcriptional feedback loop.
Kowalska E, Ripperger JA, Muheim C, Maier B, Kurihara Y, Fox AH, Kramer A, Brown SA. Kowalska E, et al. Mol Cell Biol. 2012 Nov;32(22):4585-94. doi: 10.1128/MCB.00334-12. Epub 2012 Sep 10. Mol Cell Biol. 2012. PMID: 22966205 Free PMC article. - Emerging roles for long noncoding RNAs in skeletal biology and disease.
Huynh NP, Anderson BA, Guilak F, McAlinden A. Huynh NP, et al. Connect Tissue Res. 2017 Jan;58(1):116-141. doi: 10.1080/03008207.2016.1194406. Epub 2016 Jun 2. Connect Tissue Res. 2017. PMID: 27254479 Free PMC article. Review.
References
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. - PubMed
- Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous