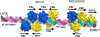The minimum replication origin of merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication - PubMed (original) (raw)
The minimum replication origin of merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication
Hyun Jin Kwun et al. J Virol. 2009 Dec.
Abstract
Merkel cell polyomavirus (MCV) is a recently discovered human polyomavirus causing the majority of human Merkel cell carcinomas. We mapped a 71-bp minimal MCV replication core origin sufficient for initiating eukaryotic DNA replication in the presence of wild-type MCV large T protein (LT). The origin includes a poly(T)-rich tract and eight variably oriented, GAGGC-like pentanucleotide sequences (PS) that serve as LT recognition sites. Mutation analysis shows that only four of the eight PS are required for origin replication. A single point mutation in one origin PS from a naturally occurring, tumor-derived virus reduces LT assembly on the origin and eliminates viral DNA replication. Tumor-derived LT having mutations truncating either the origin-binding domain or the helicase domain also prevent LT-origin assembly. Optimal MCV replication requires coexpression of MCV small T protein (sT), together with LT. An intact DnaJ domain on the LT is required for replication but is dispensable on the sT. In contrast, PP2A targeting by sT is required for enhanced replication. The MCV origin provides a novel model for eukaryotic replication from a defined DNA element and illustrates the selective pressure within tumors to abrogate independent MCV replication.
Figures
FIG. 1.
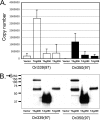
Mapping of the MCV core origin. (A) Comparison of the origin sequences among polyomavirus family. The straight and dotted arrow heads indicate complete (GAGGC) or incomplete pentanucleotide sequences (GXGGC and GAGXC) serving for LT binding sites, respectively. The origin of SV40 subgroup (GenBank accession no. EF59667.1) consists of highly conserved region, including the AT-rich tract on the late gene side, site II GAGGC repeats, the early palindrome (EP), and early enhancer region on the early gene side. The MCV origin (GenBank accession no. EU375804) is closely related to the origin of Py (GenBank accession no. J02288). MCV origin constructs of different lengths (589 to 69 bp) were cloned into pCR2.1 vector (Invitrogen) to identify the core origin sequence. (B) Deletion analysis of MCV origin. The origin constructs were cotransfected into 293 cells with either TAg206.wt or empty vector as a negative control, and the replication from the MCV origin was analyzed by Southern blotting. The autoradiogram was quantified by using ImageQuant software (GE Healthcare). The DpnI-sensitive DNA band was used as an internal standard for the amount of input DNA.
FIG. 2.
Definition of MCV minimal core origin. Single-base-pair deletions from both sides of the origin sequence (late, nucleotide 5356; early, nucleotide 48) were performed to define a minimal core origin. The expression level of TAg was observed by Western blotting (data not shown), and an autoradiogram of replication was quantified as in Fig. 1B (middle panel).
FIG. 3.
Pentanucleotide requirements for MCV origin replication. (A) Mutation of each pentanucleotide in the 97-bp origin was generated by PCR-based direct mutagenesis (AT to GC) to define the essential pentanucleotides for MCV origin replication. (B) Replication efficiency was examined by Southern blotting.
FIG. 4.
Single mutational analysis of pentanucleotides. (A) Each sequence of pentanucleotides 1, 2, 3, 7, and 8 was mutated by AT-to-GC substitutions to disrupt the TAg binding site. To avoid reconstitution of the T-antigen binding site [5′-G(A/G)GGC-3′] by mutation, AG-to-T substitutions were introduced in S7-2, S8-2, S2-2, and S3-2. (B) Southern blot results demonstrate that single base mutations in pentanucleotides 1, 2, and 7 can abolish replication efficiently, indicating that these pentanucleotides sites are critical for MCV origin replication as a functional unit.
FIG. 5.
ChIP analysis. (A) The binding efficiency of both non-tumor- and tumor-derived TAg to MCV origin in vivo was examined by using a ChIP assay. The 97-bp origin-containing plasmid [Ori339(97) or Ori350(97)] and the TAg constructs from nontumor (TAg206.wt) or tumor (TAg350 and TAg339) tissues (50) were cotransfected into 293 cells, and the binding of LT to the origin was investigated by quantitative PCR. All experiments were repeated at least three times. Error bars indicate the standard deviations. (B) The relative expression levels of wild-type and mutant LT proteins are demonstrated by Western blotting.
FIG. 6.
Intrinsic activities of T antigens that influence MCV replication. (A) Alternative spliced products (T1 to T4) of the early region give rise to LT, sT, and 57kT. Arrows indicate the positions of siRNAs, siT, and sisT. siT targets all TAg transcripts, while sisT targets only sT transcripts. An asterisk indicates the epitope site for CM8E6, and a star indicates the epitope site of CM2B4 antibodies. These antibodies detect LT/57kT/sT or LT/57kT proteins, respectively. The predicted MCV LT gene sequence retains all major conserved features of other polyomavirus LTs, including DnaJ, Rb-targeting, origin-binding, and helicase/ATPase domains. (B) sT expression significantly contributes to the efficiency of origin replication. LT, 57kT, and sT expression plasmids were cotransfected in various combinations into 293 cell. For sT expression, a green fluorescent protein (GFP) fusion construct was used. Western blotting was performed to monitor the expression level of each transcript product using V5 antibody (Invitrogen) for LT and 57kT and anti-GFP (SantaCruz) antibody for sT. A representative experiment out of three repeats is shown. (C) siRNA knockdown of LT, 57kT, and/or sT expression confirms the contribution of sT to efficient origin replication (D). A nonsilencing siRNA (siCtrl) was used as a control. Three specific bands (120, 57, and 19 kDa) are detected by Western blotting with CM8E6 antibody in 293 cells transfected with genomic TAg expression constructs (TAg206.wt) (C, right panel). CM2B4 detected both ∼120- and ∼57-kDa bands corresponding to the full-length LT and 57kT protein products, respectively (49) (C, left panel). (E) To examine the interaction between TAg and Hsc70 and its role in origin replication, Myc-tagged Hsc70 construct (pCMV-myc-Hsc70) was transfected into 293 cells with either V5-tagged wild-type TAg (TAg206.wt) or Hsc70 mutant TAg (D44H, D44N). Lysates were immunoprecipitated with anti-myc or anti-V5 antibody and immunoblotted with anti-V5 or anti-myc antibody in a reciprocal way, respectively. The LacZ gene-expressing vector, pcDNA6/V5-His/LacZ (Invitrogen), was used as a negative control for binding and 2% of the lysate was used for input control. (F) Hsc70 binding to TAg promotes efficient viral replication. For the replication assay, TAg206.wt or mutants (Hsc70 binding mutant, D44N; Rb binding mutant, LXCXK; Rb/Hsc70 binding mutant, LXCXK/D44N) were cotransfected with MCV origin plasmid [Ori339(97)] into 293 cells. Replication efficiency was measured and quantified by using ImageQuant (GE Healthcare).
FIG. 7.
Model of the MCV OBDs on the MCV origin. A model of the MCV OBD was placed on each of the putative binding sites. OBDs positioned at the required PS1, PS2, PS4, and PS7 are shown in blue. The remainder are yellow. The GAGGC sequences are shown in cyan. The AT-rich tracts of DNA that flank site 1/2 are shown in light magenta.
Similar articles
- T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus.
Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. Shuda M, et al. Proc Natl Acad Sci U S A. 2008 Oct 21;105(42):16272-7. doi: 10.1073/pnas.0806526105. Epub 2008 Sep 23. Proc Natl Acad Sci U S A. 2008. PMID: 18812503 Free PMC article. - [Merkel cell polyomavirus and Merkel cell carcinoma].
Nakamura T, Katano H. Nakamura T, et al. Uirusu. 2009 Jun;59(1):37-42. doi: 10.2222/jsv.59.37. Uirusu. 2009. PMID: 19927987 Review. Japanese. - Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin.
Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. Harrison CJ, et al. J Mol Biol. 2011 Jun 17;409(4):529-42. doi: 10.1016/j.jmb.2011.03.051. Epub 2011 Apr 9. J Mol Biol. 2011. PMID: 21501625 Free PMC article. - Unlicensed origin DNA melting by MCV and SV40 polyomavirus LT proteins is independent of ATP-dependent helicase activity.
Wan L, Toland S, Robinson-McCarthy LR, Lee N, Schaich MA, Hengel SR, Li X, Bernstein KA, Van Houten B, Chang Y, Moore PS. Wan L, et al. Proc Natl Acad Sci U S A. 2023 Jul 25;120(30):e2308010120. doi: 10.1073/pnas.2308010120. Epub 2023 Jul 17. Proc Natl Acad Sci U S A. 2023. PMID: 37459531 Free PMC article. - Large T and small T antigens of Merkel cell polyomavirus.
Wendzicki JA, Moore PS, Chang Y. Wendzicki JA, et al. Curr Opin Virol. 2015 Apr;11:38-43. doi: 10.1016/j.coviro.2015.01.009. Epub 2015 Feb 11. Curr Opin Virol. 2015. PMID: 25681708 Free PMC article. Review.
Cited by
- Regulation of Polyomavirus Transcription by Viral and Cellular Factors.
Yang JF, You J. Yang JF, et al. Viruses. 2020 Sep 24;12(10):1072. doi: 10.3390/v12101072. Viruses. 2020. PMID: 32987952 Free PMC article. Review. - Merkel Cell Polyomavirus and Merkel Cell Carcinoma.
Pietropaolo V, Prezioso C, Moens U. Pietropaolo V, et al. Cancers (Basel). 2020 Jul 3;12(7):1774. doi: 10.3390/cancers12071774. Cancers (Basel). 2020. PMID: 32635198 Free PMC article. Review. - Combining DNA Damage Induction with BCL-2 Inhibition to Enhance Merkel Cell Carcinoma Cytotoxicity.
Liu W, Krump NA, Herlyn M, You J. Liu W, et al. Biology (Basel). 2020 Feb 19;9(2):35. doi: 10.3390/biology9020035. Biology (Basel). 2020. PMID: 32093022 Free PMC article. - The HSP70 modulator MAL3-101 inhibits Merkel cell carcinoma.
Adam C, Baeurle A, Brodsky JL, Wipf P, Schrama D, Becker JC, Houben R. Adam C, et al. PLoS One. 2014 Apr 2;9(4):e92041. doi: 10.1371/journal.pone.0092041. eCollection 2014. PLoS One. 2014. PMID: 24694787 Free PMC article. - Protein-mediated viral latency is a novel mechanism for Merkel cell polyomavirus persistence.
Kwun HJ, Chang Y, Moore PS. Kwun HJ, et al. Proc Natl Acad Sci U S A. 2017 May 16;114(20):E4040-E4047. doi: 10.1073/pnas.1703879114. Epub 2017 May 1. Proc Natl Acad Sci U S A. 2017. PMID: 28461484 Free PMC article.
References
- Benbow, R. M., J. Zhao, and D. D. Larson. 1992. On the nature of origins of DNA replication in eukaryotes. Bioessays 14:661-670. - PubMed
- Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. Zalvide, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098-1110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 AI082496/AI/NIAID NIH HHS/United States
- R01 CA136363/CA/NCI NIH HHS/United States
- R33 CA120726/CA/NCI NIH HHS/United States
- CA136363/CA/NCI NIH HHS/United States
- CA120726/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources