Gel electrophoresis assays for analyzing DNA double-strand breaks in Saccharomyces cerevisiae at various spatial resolutions - PubMed (original) (raw)
Review
Gel electrophoresis assays for analyzing DNA double-strand breaks in Saccharomyces cerevisiae at various spatial resolutions
Hajime Murakami et al. Methods Mol Biol. 2009.
Abstract
Meiotic recombination is triggered by programmed DNA double-strand breaks (DSBs), which are catalyzed by Spo11 protein in a type II topoisomerase-like manner. Meiotic DSBs can be detected directly using physical assays (gel electrophoresis, Southern blotting, and indirect end-labeling) applied to samples of genomic DNA from sporulating cultures of budding and fission yeast. Such assays are extremely useful for quantifying and characterizing many aspects of the initiation of meiotic recombination, including the timing of DSB formation relative to other events, the distribution of DSBs across the genome, and the influence on DSB formation of mutations in recombination factors and other gene products. By varying the type of gel electrophoresis and other parameters, the spatial resolution of DSB analysis can range from single nucleotides up to whole yeast chromosomes.
Figures
Fig. 1
The procedure of DSB mapping at four different resolutions. Left: General procedure of DSB mapping. Genomic DNA is digested with restriction enzyme(s) to yield appropriately sized DNA fragments (for chromosome-level mapping, this step is skipped). The DNA fragments are separated by electrophoresis on an appropriate gel according to the size of the fragment, then detected by Southern blot hybridization. Right: Comparison of methods and resolution of chromosome, medium, high, and nucleotide level resolution mapping.
Fig. 2
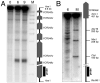
Chromosome level mapping. Cells from a s_ae2_Δ strain (MJL 2305) (A) or a wild-type strain (MJL 1071) (B) were harvested at the indicated times in meiosis and processed for genomic DNA plug preparation and pulsed-field gel electrophoresis as described in the text. After Southern blotting, the membrane was probed with a chromosome III left end fragment (CHA1), revealing, in addition to the unbroken chromosome III (340 kb), the DSB fragments of chromosome III produced during meiosis. In (A), a 14 cm wide by 21 cm long gel was used, and electrophoresis time was for 46 h. In (B), a 21 cm wide by 14 cm long gel was used and electrophoresis time was 30.5 h. The size standard (M) is a Hind III digest of bacteriophage λ DNA and λ DNA concatamers. (B) is adapted with permission from (8).
Fig. 3
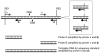
Medium and high resolution mapping of DSB in a rad50S strain. (A) Medium resolution mapping of DSBs around YCR048w. Numbers above the panel indicate the time (in h) after transfer to SPM. The lane marked M includes 200 ng of λ _Bst_E II digest. Genomic DNA was digested with Ase I. The parental fragment is 9.6 kb. Both the genomic DNA and the marker were separated on a 0.8 % agarose gel and detected using a radiolabeled probe containing 100 ng of the probe DNA fragment and 0.1 ng of λ DNA. (B) High resolution mapping of DSBs in the YCR048w promoter region. DSBs, which appeared as a single band in normal resolution mapping, are observed as multiple break sites when analyzed at this resolution. Genomic DNA extracted from meiotic cells (6 h) was digested with Xmn I and _Bsu_36 I. For the molecular weight marker, genomic DNA purified from 0 h cells was digested with Xmn I and _Bsu_36 I. Aliquots (1% of the digested DNA each) were further digested with either _Alw_N I, Msc I or Cla I. After heat inactivation of the restriction enzymes, the differently digested DNA fragments were mixed together and loaded in the lane marked M. These DNA fragments were separated on a 6% polyacrylamide gel containing 8 M urea and detected by Southern blotting and indirect end labeling.
Fig. 4
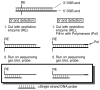
Primers required for nucleotide level mapping. For nucleotide resolution mapping, genomic DNA is digested with an appropriate restriction enzyme (RE1 or RE2), which cleaves 150–200 bp from DSB sites. Four primers (a, b, c, and d) are used for following procedures: to amplify probes A and B and a template DNA for sequence standards; to prepare strand-specific probe (primers b and c); to prepare sequence standards (primers a and d). Primers a and d are designed to be suitable to the shape of restriction enzyme cut as illustrated in the box on the right.
Fig. 5
Overview of procedure for nucleotide level resolution mapping of 5′ and 3′ DSB ends. Genomic DNA is purified from meiotic cell and digested with an appropriate restriction enzyme (RE). After the separation on a sequencing gel and Southern blotting, 3′ DSB ends are detected with a strand-specific DNA probe. For the indirect detection and mapping of 5′ ends, 3′ ends are filled-in with DNA polymerase to match the size of the 5′ ends.
Fig. 6
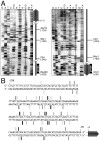
Nucleotide level resolution mapping of DSB at YCR048w promoter region. (A) Examples of sequencing gel mapping of DSBs. Images of Southern blots probed with two different probes are shown. Samples were digested with _Bsu_36 I (left) or Xmn I (right) and probed with single-stranded DNA in the vicinity of each restriction enzyme site (illustrated on the right of each panel; see also Fig. 4). Lanes marked 5′ and 3′ contain separate DSB end-mapping samples, prepared using genomic DNA purified from cells 9 h after transfer to SPM. Lanes marked G, A, T, and C are nucleotide sequence standards. Lanes marked GC and AT are sequencing standard containing pooled G+C or A+T standards, respectively. All of the sequence standards contain additional restriction-digested genomic DNA (from a 0 h culture) so that the amount of total genomic DNA is the same as in the DSB sample lanes. Lanes marked M contain DNA from 0 h cells, digested with either _Bsu_36 I (left) or Xmn I (right), plus 1% of DNA subjected to secondary digest with either _Alw_N I, Msc I, or Cla I (see Note 12). Numbers correspond to positions relative to the translation start of YCR048w. (B) Location of DSBs in the YCR048w promoter region. The vertical bars represent DSB locations, defined by mapping of DSB 5′ ends using probes on both sides of the DSB hotspot. Thickness of the bars provides a semi-quantitative representation of the signal strength of each DSB band on the Southern blots.
Similar articles
- Sequencing Spo11 Oligonucleotides for Mapping Meiotic DNA Double-Strand Breaks in Yeast.
Lam I, Mohibullah N, Keeney S. Lam I, et al. Methods Mol Biol. 2017;1471:51-98. doi: 10.1007/978-1-4939-6340-9_3. Methods Mol Biol. 2017. PMID: 28349390 - Locally, meiotic double-strand breaks targeted by Gal4BD-Spo11 occur at discrete sites with a sequence preference.
Murakami H, Nicolas A. Murakami H, et al. Mol Cell Biol. 2009 Jul;29(13):3500-16. doi: 10.1128/MCB.00088-09. Epub 2009 Apr 20. Mol Cell Biol. 2009. PMID: 19380488 Free PMC article. - Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae.
Buhler C, Borde V, Lichten M. Buhler C, et al. PLoS Biol. 2007 Dec;5(12):e324. doi: 10.1371/journal.pbio.0050324. PLoS Biol. 2007. PMID: 18076285 Free PMC article. - Genome-wide mapping of meiotic DNA double-strand breaks in Saccharomyces cerevisiae.
Buhler C, Shroff R, Lichten M. Buhler C, et al. Methods Mol Biol. 2009;557:143-64. doi: 10.1007/978-1-59745-527-5_10. Methods Mol Biol. 2009. PMID: 19799181 Review. - Modulating and targeting meiotic double-strand breaks in Saccharomyces cerevisiae.
Nicolas A. Nicolas A. Methods Mol Biol. 2009;557:27-33. doi: 10.1007/978-1-59745-527-5_3. Methods Mol Biol. 2009. PMID: 19799174 Review.
Cited by
- Smc5/6 coordinates formation and resolution of joint molecules with chromosome morphology to ensure meiotic divisions.
Copsey A, Tang S, Jordan PW, Blitzblau HG, Newcombe S, Chan AC, Newnham L, Li Z, Gray S, Herbert AD, Arumugam P, Hochwagen A, Hunter N, Hoffmann E. Copsey A, et al. PLoS Genet. 2013;9(12):e1004071. doi: 10.1371/journal.pgen.1004071. Epub 2013 Dec 26. PLoS Genet. 2013. PMID: 24385939 Free PMC article. - Programming sites of meiotic crossovers using Spo11 fusion proteins.
Sarno R, Vicq Y, Uematsu N, Luka M, Lapierre C, Carroll D, Bastianelli G, Serero A, Nicolas A. Sarno R, et al. Nucleic Acids Res. 2017 Nov 2;45(19):e164. doi: 10.1093/nar/gkx739. Nucleic Acids Res. 2017. PMID: 28977556 Free PMC article. - Chromosome-autonomous feedback down-regulates meiotic DNA break competence upon synaptonemal complex formation.
Mu X, Murakami H, Mohibullah N, Keeney S. Mu X, et al. Genes Dev. 2020 Dec 1;34(23-24):1605-1618. doi: 10.1101/gad.342873.120. Epub 2020 Nov 12. Genes Dev. 2020. PMID: 33184224 Free PMC article. - Regulation of the MLH1-MLH3 endonuclease in meiosis.
Cannavo E, Sanchez A, Anand R, Ranjha L, Hugener J, Adam C, Acharya A, Weyland N, Aran-Guiu X, Charbonnier JB, Hoffmann ER, Borde V, Matos J, Cejka P. Cannavo E, et al. Nature. 2020 Oct;586(7830):618-622. doi: 10.1038/s41586-020-2592-2. Epub 2020 Aug 19. Nature. 2020. PMID: 32814904 - Extensive Recombination of a Yeast Diploid Hybrid through Meiotic Reversion.
Laureau R, Loeillet S, Salinas F, Bergström A, Legoix-Né P, Liti G, Nicolas A. Laureau R, et al. PLoS Genet. 2016 Feb 1;12(2):e1005781. doi: 10.1371/journal.pgen.1005781. eCollection 2016 Feb. PLoS Genet. 2016. PMID: 26828862 Free PMC article.
References
- Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2:360–369. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases