Reassortment patterns in Swine influenza viruses - PubMed (original) (raw)
Reassortment patterns in Swine influenza viruses
Hossein Khiabanian et al. PLoS One. 2009.
Abstract
Three human influenza pandemics occurred in the twentieth century, in 1918, 1957, and 1968. Influenza pandemic strains are the results of emerging viruses from non-human reservoirs to which humans have little or no immunity. At least two of these pandemic strains, in 1957 and in 1968, were the results of reassortments between human and avian viruses. Also, many cases of swine influenza viruses have reportedly infected humans, in particular, the recent H1N1 influenza virus of swine origin, isolated in Mexico and the United States. Pigs are documented to allow productive replication of human, avian, and swine influenza viruses. Thus it has been conjectured that pigs are the "mixing vessel" that create the avian-human reassortant strains, causing the human pandemics. Hence, studying the process and patterns of viral reassortment, especially in pigs, is a key to better understanding of human influenza pandemics. In the last few years, databases containing sequences of influenza A viruses, including swine viruses, collected since 1918 from diverse geographical locations, have been developed and made publicly available. In this paper, we study an ensemble of swine influenza viruses to analyze the reassortment phenomena through several statistical techniques. The reassortment patterns in swine viruses prove to be similar to the previous results found in human viruses, both in vitro and in vivo, that the surface glycoprotein coding segments reassort most often. Moreover, we find that one of the polymerase segments (PB1), reassorted in the strains responsible for the last two human pandemics, also reassorts frequently.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
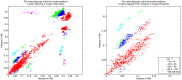
Figure 1. Pair-wise hamming distances at the third codon positions in PB2 vs. PB1.
The colors demonstrate the logarithm of the cumulative probability for the points, among which the ones with a cumulative probability of less than 10−7 indicate possible reassortment events. Left: The results from 150 strains in the dataset, where there are candidates for reassortment events in both PB2 and PB1. Right: The results when the dataset is limited to the classical H1N1 strains isolated in the 70's, 80's, and 90's, where there are distinctively more candidates for reassortment events in PB1.
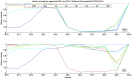
Figure 2. History overlap for segments PB1 and NP of the swine influenza strain A/Swine/Tennessee/23/1976.
NP and all the other segments except PB1 share a common recent history, whereas the recent history of PB1 is different from the other seven segments, indicating a reassortment event at PB1. The small history overlap of M1 with PB1 and NP at lower identities can be attributed to a possible slower evolutionary rate of M1 and a fork in its lineage to a line of human viruses. The fluctuations in the history overlap of NP at 99% identity are due to small number of sample points at that level of identity.
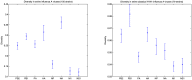
Figure 3. Diversity measurements in swine influenza viruses and the corresponding 95% bootstrap percentile confidence intervals.
Left: Considering the 150 strains in the dataset, NA, HA, and PB1 present a higher diversity than the rest. Right: When the dataset is limited to the classical H1N1 strains isolated in the 70's, 80's, and 90's, which fixes the HA and NA variations in the population, shows a higher diversity in PB1 than the rest of the segments.
Figure 4. Pair-wise Pearson correlation of the distances at the third codon positions of the viral segments.
Left: The HA, NA, and PB1 segments have the least correlation in regards to the rest of the segments. Right: When the HA and NA variations are fixed in the population by limiting the dataset to the classical H1N1 strains isolated in the 70's, 80's, and 90's, PB1 presents a distinctively lower correlation relative to the other segments.
Update of
- Reassortment patterns in Swine influenza viruses.
Khiabanian H, Trifonov V, Rabadan R. Khiabanian H, et al. PLoS Curr. 2009 Aug 21;1:RRN1008. doi: 10.1371/currents.RRN1008. PLoS Curr. 2009. PMID: 20029610 Free PMC article.
Similar articles
- Reassortment patterns in Swine influenza viruses.
Khiabanian H, Trifonov V, Rabadan R. Khiabanian H, et al. PLoS Curr. 2009 Aug 21;1:RRN1008. doi: 10.1371/currents.RRN1008. PLoS Curr. 2009. PMID: 20029610 Free PMC article. - Swine Influenza Virus PA and Neuraminidase Gene Reassortment into Human H1N1 Influenza Virus Is Associated with an Altered Pathogenic Phenotype Linked to Increased MIP-2 Expression.
Dlugolenski D, Jones L, Howerth E, Wentworth D, Tompkins SM, Tripp RA. Dlugolenski D, et al. J Virol. 2015 May;89(10):5651-67. doi: 10.1128/JVI.00087-15. Epub 2015 Mar 11. J Virol. 2015. PMID: 25762737 Free PMC article. - Evolutionary genomics of the pandemic 2009 H1N1 influenza viruses (pH1N 1v).
Qu Y, Zhang R, Cui P, Song G, Duan Z, Lei F. Qu Y, et al. Virol J. 2011 May 21;8:250. doi: 10.1186/1743-422X-8-250. Virol J. 2011. PMID: 21600019 Free PMC article. - In vivo reassortment of influenza viruses.
Urbaniak K, Markowska-Daniel I. Urbaniak K, et al. Acta Biochim Pol. 2014;61(3):427-31. Epub 2014 Sep 3. Acta Biochim Pol. 2014. PMID: 25180223 Review. - Isolation and genetic characterization of avian-like H1N1 and novel ressortant H1N2 influenza viruses from pigs in China.
Yu H, Zhang PC, Zhou YJ, Li GX, Pan J, Yan LP, Shi XX, Liu HL, Tong GZ. Yu H, et al. Biochem Biophys Res Commun. 2009 Aug 21;386(2):278-83. doi: 10.1016/j.bbrc.2009.05.056. Epub 2009 May 19. Biochem Biophys Res Commun. 2009. PMID: 19460353 Review.
Cited by
- Unaltered influenza disease outcomes in swine prophylactically treated with α-galactosylceramide.
Gu W, Madrid DMD, Yang G, Artiaga BL, Loeb JC, Castleman WL, Richt JA, Lednicky JA, Driver JP. Gu W, et al. Dev Comp Immunol. 2021 Jan;114:103843. doi: 10.1016/j.dci.2020.103843. Epub 2020 Aug 29. Dev Comp Immunol. 2021. PMID: 32871161 Free PMC article. - Reassortment Networks and the evolution of pandemic H1N1 swine-origin influenza.
Bokhari SH, Pomeroy LW, Janies DA. Bokhari SH, et al. IEEE/ACM Trans Comput Biol Bioinform. 2012 Jan-Feb;9(1):214-27. doi: 10.1109/TCBB.2011.95. IEEE/ACM Trans Comput Biol Bioinform. 2012. PMID: 22076498 Free PMC article. - Detection of H1 Swine Influenza A Virus Antibodies in Human Serum Samples by Age Group1.
Vandoorn E, Leroux-Roels I, Leroux-Roels G, Parys A, Vincent A, Van Reeth K. Vandoorn E, et al. Emerg Infect Dis. 2020 Sep;26(9):2118-2128. doi: 10.3201/eid2609.191796. Emerg Infect Dis. 2020. PMID: 32818398 Free PMC article. - Viral reassortment as an information exchange between viral segments.
Greenbaum BD, Li OT, Poon LL, Levine AJ, Rabadan R. Greenbaum BD, et al. Proc Natl Acad Sci U S A. 2012 Feb 28;109(9):3341-6. doi: 10.1073/pnas.1113300109. Epub 2012 Feb 13. Proc Natl Acad Sci U S A. 2012. PMID: 22331898 Free PMC article. - Progress and Challenge in Computational Identification of Influenza Virus Reassortment.
Ding X, Qin L, Meng J, Peng Y, Wu A, Jiang T. Ding X, et al. Virol Sin. 2021 Dec;36(6):1273-1283. doi: 10.1007/s12250-021-00392-w. Epub 2021 May 26. Virol Sin. 2021. PMID: 34037948 Free PMC article. Review.
References
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, et al. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437(7060):889–93. - PubMed
- Lindstrom SE, Cox NJ, Klimov A. Genetic analysis of human H2N2 and early H3N2 influenza viruses, 1957–1972: evidence for genetic divergence and multiple reassortment events. Virology. 2004;328(1):101–19. - PubMed
- Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87(1):13–20. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous