edgeR: a Bioconductor package for differential expression analysis of digital gene expression data - PubMed (original) (raw)
edgeR: a Bioconductor package for differential expression analysis of digital gene expression data
Mark D Robinson et al. Bioinformatics. 2010.
Abstract
Summary: It is expected that emerging digital gene expression (DGE) technologies will overtake microarray technologies in the near future for many functional genomics applications. One of the fundamental data analysis tasks, especially for gene expression studies, involves determining whether there is evidence that counts for a transcript or exon are significantly different across experimental conditions. edgeR is a Bioconductor software package for examining differential expression of replicated count data. An overdispersed Poisson model is used to account for both biological and technical variability. Empirical Bayes methods are used to moderate the degree of overdispersion across transcripts, improving the reliability of inference. The methodology can be used even with the most minimal levels of replication, provided at least one phenotype or experimental condition is replicated. The software may have other applications beyond sequencing data, such as proteome peptide count data.
Availability: The package is freely available under the LGPL licence from the Bioconductor web site (http://bioconductor.org).
Figures
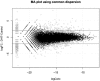
Fig. 1.
DGE data can be visualized as ‘MA’ plots (log ratio versus abundance), just as with microarray data where each dot represents a gene. This plot shows RNA-seq gene expression for DHT-stimulated versus Control LNCaP cells, as described in Li et al. (2008). The smear of points on the left side signifies that genes were observed in only one group of replicate samples and the points marked ‘×’ denote the top 500 differentially expressed genes.
Similar articles
- Codelink: an R package for analysis of GE healthcare gene expression bioarrays.
Diez D, Alvarez R, Dopazo A. Diez D, et al. Bioinformatics. 2007 May 1;23(9):1168-9. doi: 10.1093/bioinformatics/btm072. Epub 2007 Mar 7. Bioinformatics. 2007. PMID: 17344240 - oneChannelGUI: a graphical interface to Bioconductor tools, designed for life scientists who are not familiar with R language.
Sanges R, Cordero F, Calogero RA. Sanges R, et al. Bioinformatics. 2007 Dec 15;23(24):3406-8. doi: 10.1093/bioinformatics/btm469. Epub 2007 Sep 17. Bioinformatics. 2007. PMID: 17875544 - SScore: an R package for detecting differential gene expression without gene expression summaries.
Kennedy RE, Kerns RT, Kong X, Archer KJ, Miles MF. Kennedy RE, et al. Bioinformatics. 2006 May 15;22(10):1272-4. doi: 10.1093/bioinformatics/btl108. Epub 2006 Mar 30. Bioinformatics. 2006. PMID: 16574698 - Pre-processing of microarray data and analysis of differential expression.
Durinck S. Durinck S. Methods Mol Biol. 2008;452:89-110. doi: 10.1007/978-1-60327-159-2_4. Methods Mol Biol. 2008. PMID: 18563370 Review. - Open source software for the analysis of microarray data.
Dudoit S, Gentleman RC, Quackenbush J. Dudoit S, et al. Biotechniques. 2003 Mar;Suppl:45-51. Biotechniques. 2003. PMID: 12664684 Review.
Cited by
- The restricted N-glycome of neurons is programmed during differentiation.
Kiwimagi K, Noel M, Cetinbas M, Sadreyev RI, Wang L, Smoller JW, Cummings RD, Weiss R, Mealer RG. Kiwimagi K, et al. bioRxiv [Preprint]. 2024 Oct 17:2024.10.15.618477. doi: 10.1101/2024.10.15.618477. bioRxiv. 2024. PMID: 39463965 Free PMC article. Preprint. - Functional Optimization in Distinct Tissues and Conditions Constrains the Rate of Protein Evolution.
Usmanova DR, Plata G, Vitkup D. Usmanova DR, et al. Mol Biol Evol. 2024 Oct 4;41(10):msae200. doi: 10.1093/molbev/msae200. Mol Biol Evol. 2024. PMID: 39431545 Free PMC article. - Regulation of Sex-biased Gene Expression by the Ancestral X-Y Chromosomal Gene Pair Kdm5c-Kdm5d.
Malcore RM, Samanta MK, Kalantry S, Iwase S. Malcore RM, et al. bioRxiv [Preprint]. 2024 Oct 27:2024.10.24.620066. doi: 10.1101/2024.10.24.620066. bioRxiv. 2024. PMID: 39484414 Free PMC article. Preprint. - Identification of a Novel Four-Gene Signature Correlated With the Prognosis of Patients With Hepatocellular Carcinoma: A Comprehensive Analysis.
Zhu W, Ru L, Ma Z. Zhu W, et al. Front Oncol. 2021 Mar 12;11:626654. doi: 10.3389/fonc.2021.626654. eCollection 2021. Front Oncol. 2021. PMID: 33777771 Free PMC article. - Comprehensive at-arrival transcriptomic analysis of post-weaned beef cattle uncovers type I interferon and antiviral mechanisms associated with bovine respiratory disease mortality.
Scott MA, Woolums AR, Swiderski CE, Perkins AD, Nanduri B, Smith DR, Karisch BB, Epperson WB, Blanton JR. Scott MA, et al. PLoS One. 2021 Apr 26;16(4):e0250758. doi: 10.1371/journal.pone.0250758. eCollection 2021. PLoS One. 2021. PMID: 33901263 Free PMC article.
References
- Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–2887. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources