Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats - PubMed (original) (raw)
Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats
Maya K Sethi et al. J Biol Chem. 2010.
Abstract
The epidermal growth factor repeats of the Notch receptor are extensively glycosylated with three different O-glycans. O-Fucosylation and elongation by the glycosyltransferase Fringe have been well studied and shown to be essential for proper Notch signaling. In contrast, biosynthesis of O-glucose and O-N-acetylglucosamine is less well understood. Recently, the isolation of the Drosophila mutant rumi has shown that absence of O-glucose impairs Notch function. O-Glucose is further extended by two contiguous alpha1,3-linked xylose residues. We have identified two enzymes of the human glycosyltransferase 8 family, now named GXYLT1 and GXYLT2 (glucoside xylosyltransferase), as UDP-d-xylose:beta-d-glucoside alpha1,3-d-xylosyltransferases adding the first xylose. The enzymes are specific for beta-glucose-terminating acceptors and UDP-xylose as donor substrate. Generation of the alpha1,3-linkage was confirmed by nuclear magnetic resonance. Activity on a natural acceptor could be shown by in vitro xylosylation of a Notch fragment expressed in a UDP-xylose-deficient cell line and in vivo by co-expression of the enzymes and the Notch fragment in insect cells followed by mass spectrometric analysis of peptide fragments.
Figures
FIGURE 1.
Sequence alignment. The complete amino acid sequence of four human GLT8 family members is aligned with the N-terminal catalytic domain of UGGT starting at amino acid 1243. Sequence conservation is shown when three out of five amino acids are identical. Putative transmembrane domains are shown shaded, and the conserved D_X_D motif is underlined.
FIGURE 2.
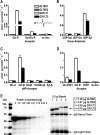
In vitro enzymatic activity. A, activity of GLT8D1, GLT8D2, GXYLT1, and GXYLT2 with acceptors mimicking the two naturally occurring xylosyltransferase substrates on EGF domains using UDP-Xyl as donor substrate. no Acc., no acceptor. B, donor substrate specificity using Glc-R as substrate. C, acceptor substrate specificity of GXYLT1 and GXYLT2 using para-nitrophenol (pNP)-linked monosaccharides as acceptor substrates. D, glucosylated EGF1 of factor VII when compared with unglucosylated EGF1 and Glc-R as acceptor, now measured at 2 μ
m
acceptor concentration (when compared with 100 μ
m
in A–C) to adapt to the low availability of factor VII EGF1. E, quantification of enzyme bound to 0.5 μl of IgG beads using a protein A standard and Coomassie Blue staining, of which 10 μl was used per assay in A–D.
FIGURE 3.
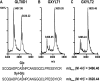
Xylosyltransferase activity using Notch EGF1–5 as acceptor. EGF1–5 produced in UDP-Xyl synthase-negative cells (A) was incubated with GXYLT1 or GXYLT2 (B and C), and tryptic peptides were analyzed by liquid chromatography-electrospray ionization-TOF mass spectrometry. Shown is the _O_-glucosylated peptide of EGF4. GXYLT1 and GXYLT2 were able to extend the _O_-glucosylated peptide by one xylose but not to produce the full trisaccharide that is produced in wild type CHO cells (D). To confirm that the m/z values shown represent the differentially glycosylated peptides of EGF4, sequencing was done by tandem mass spectrometry and shown in
supplemental Fig. S2
. MW, molecular weight.
FIGURE 4.
In vivo xylosyltransferase activity. Shown are the MALDI-mass spectra of insect cell-produced EGF1–5 that was co-infected with GLT8D1 as a negative control (A), GXYLT1 (B), or GXYLT2 (C). The glucosylated peptide with the calculated nominal mass of 3498 Da is most dominant in A but is reduced in favor of the Xyl-Glc-peptide (3630 Da) after incubation with GXYLT1 or GXYLT2. As for Fig. 3, peptide sequencing was performed and is presented in
supplemental Fig. S2
.
Similar articles
- Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch.
Sethi MK, Buettner FF, Ashikov A, Krylov VB, Takeuchi H, Nifantiev NE, Haltiwanger RS, Gerardy-Schahn R, Bakker H. Sethi MK, et al. J Biol Chem. 2012 Jan 20;287(4):2739-48. doi: 10.1074/jbc.M111.302406. Epub 2011 Nov 23. J Biol Chem. 2012. PMID: 22117070 Free PMC article. - Site-specific O-glucosylation of the epidermal growth factor-like (EGF) repeats of notch: efficiency of glycosylation is affected by proper folding and amino acid sequence of individual EGF repeats.
Takeuchi H, Kantharia J, Sethi MK, Bakker H, Haltiwanger RS. Takeuchi H, et al. J Biol Chem. 2012 Oct 5;287(41):33934-44. doi: 10.1074/jbc.M112.401315. Epub 2012 Aug 7. J Biol Chem. 2012. PMID: 22872643 Free PMC article. - Negative regulation of notch signaling by xylose.
Lee TV, Sethi MK, Leonardi J, Rana NA, Buettner FF, Haltiwanger RS, Bakker H, Jafar-Nejad H. Lee TV, et al. PLoS Genet. 2013 Jun;9(6):e1003547. doi: 10.1371/journal.pgen.1003547. Epub 2013 Jun 6. PLoS Genet. 2013. PMID: 23754965 Free PMC article. - Significance of glycosylation in Notch signaling.
Takeuchi H, Haltiwanger RS. Takeuchi H, et al. Biochem Biophys Res Commun. 2014 Oct 17;453(2):235-42. doi: 10.1016/j.bbrc.2014.05.115. Epub 2014 Jun 6. Biochem Biophys Res Commun. 2014. PMID: 24909690 Free PMC article. Review. - Effects of Notch glycosylation on health and diseases.
Urata Y, Takeuchi H. Urata Y, et al. Dev Growth Differ. 2020 Jan;62(1):35-48. doi: 10.1111/dgd.12643. Epub 2019 Dec 30. Dev Growth Differ. 2020. PMID: 31886522 Review.
Cited by
- Significant Roles of Notch _O_-Glycosylation in Cancer.
Wang W, Okajima T, Takeuchi H. Wang W, et al. Molecules. 2022 Mar 9;27(6):1783. doi: 10.3390/molecules27061783. Molecules. 2022. PMID: 35335147 Free PMC article. Review. - Multiple roles for O-glycans in Notch signalling.
Varshney S, Stanley P. Varshney S, et al. FEBS Lett. 2018 Dec;592(23):3819-3834. doi: 10.1002/1873-3468.13251. Epub 2018 Nov 28. FEBS Lett. 2018. PMID: 30207383 Free PMC article. Review. - Fluorescence-Polarization-Based Assaying of Lysozyme with Chitooligosaccharide Tracers.
Mukhametova LI, Zherdev DO, Kuznetsov AN, Yudina ON, Tsvetkov YE, Eremin SA, Krylov VB, Nifantiev NE. Mukhametova LI, et al. Biomolecules. 2024 Jan 31;14(2):170. doi: 10.3390/biom14020170. Biomolecules. 2024. PMID: 38397407 Free PMC article. - Identification of Candidate Male-Reproduction-Related Genes from the Testis and Androgenic Gland of Macrobrachium nipponense, Regulated by PDHE1, through Transcriptome Profiling Analysis.
Jin S, Xiong Y, Zhang W, Qiao H, Wu Y, Jiang S, Fu H. Jin S, et al. Int J Mol Sci. 2024 Feb 5;25(3):1940. doi: 10.3390/ijms25031940. Int J Mol Sci. 2024. PMID: 38339218 Free PMC article. - Mucin-Type O-Glycosylation in Invertebrates.
Staudacher E. Staudacher E. Molecules. 2015 Jun 9;20(6):10622-40. doi: 10.3390/molecules200610622. Molecules. 2015. PMID: 26065637 Free PMC article. Review.
References
- Moloney D. J., Shair L. H., Lu F. M., Xia J., Locke R., Matta K. L., Haltiwanger R. S. (2000) J. Biol. Chem. 275, 9604–9611 - PubMed
- Matsuura A., Ito M., Sakaidani Y., Kondo T., Murakami K., Furukawa K., Nadano D., Matsuda T., Okajima T. (2008) J. Biol. Chem. 283, 35486–35495 - PubMed
- Okamura Y., Saga Y. (2008) Mech. Dev. 125, 663–673 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases