Evolution of MRSA during hospital transmission and intercontinental spread - PubMed (original) (raw)
. 2010 Jan 22;327(5964):469-74.
doi: 10.1126/science.1182395.
Edward J Feil, Matthew T G Holden, Michael A Quail, Emma K Nickerson, Narisara Chantratita, Susana Gardete, Ana Tavares, Nick Day, Jodi A Lindsay, Jonathan D Edgeworth, Hermínia de Lencastre, Julian Parkhill, Sharon J Peacock, Stephen D Bentley
Affiliations
- PMID: 20093474
- PMCID: PMC2821690
- DOI: 10.1126/science.1182395
Evolution of MRSA during hospital transmission and intercontinental spread
Simon R Harris et al. Science. 2010.
Abstract
Current methods for differentiating isolates of predominant lineages of pathogenic bacteria often do not provide sufficient resolution to define precise relationships. Here, we describe a high-throughput genomics approach that provides a high-resolution view of the epidemiology and microevolution of a dominant strain of methicillin-resistant Staphylococcus aureus (MRSA). This approach reveals the global geographic structure within the lineage, its intercontinental transmission through four decades, and the potential to trace person-to-person transmission within a hospital environment. The ability to interrogate and resolve bacterial populations is applicable to a range of infectious diseases, as well as microbial ecology.
Figures
Fig. 1
Phylogenetic evidence for intercontinental spread and hospital transmission of ST239 isolates. Maximum likelihood phylogenetic tree based on core genome SNPs of ST239 isolates, annotated with the country and year of isolation. The continental origin of each isolate is indicated by the color of the isolate name: blue, Asia; black, North America; green, South America; red, Europe; and yellow, Australasia. Bootstrap values are shown below each branch, with a star representing 100% bootstrap support. The scale bar represents substitutions per SNP site. A cladogram of the Thai clade is displayed for greater resolution with bootstrap values (above the branch), number of distinguishing SNPs (below the branch), and isolates labeled with date of isolation, where known.
Fig. 2
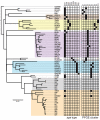
Comparison of phylogeny with traditional typing techniques. Maximum likelihood phylogenetic tree based on core genome SNPs of ST239 isolates, annotated with spa typing databased on the RIDOM scheme (27), and PFGE typing databased on BioNumerics (version 4.0, Applied Maths, Ghent, Belgium) clustering (excluding the Thai hospital isolates and USA300, which had not been typed). The most common spa type was t037, which accounted for all but one of the isolates corresponding to the South American clade but was also represented among a scattering of isolates from Europe and Asia, suggesting that t037 represents the ancestral ST239 spa type (the plesiomorphic state). Solid boxes in the appropriate column indicate the respective spa type (left grid) and PFGE cluster (right grid) of the strain. Major clades in the tree are shaded for clarity.
Similar articles
- Demography and Intercontinental Spread of the USA300 Community-Acquired Methicillin-Resistant Staphylococcus aureus Lineage.
Glaser P, Martins-Simões P, Villain A, Barbier M, Tristan A, Bouchier C, Ma L, Bes M, Laurent F, Guillemot D, Wirth T, Vandenesch F. Glaser P, et al. mBio. 2016 Feb 16;7(1):e02183-15. doi: 10.1128/mBio.02183-15. mBio. 2016. PMID: 26884428 Free PMC article. - Next-Generation Sequencing Confirms Presumed Nosocomial Transmission of Livestock-Associated Methicillin-Resistant Staphylococcus aureus in the Netherlands.
Bosch T, Witteveen S, Haenen A, Landman F, Schouls LM. Bosch T, et al. Appl Environ Microbiol. 2016 Jun 30;82(14):4081-4089. doi: 10.1128/AEM.00773-16. Print 2016 Jul 15. Appl Environ Microbiol. 2016. PMID: 27129960 Free PMC article. - Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with Methicillin-resistant S. aureus hospital-acquired infection in the United States.
Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MT, Godfrey P, Palmer KL, Bodi K, Mongodin EF, Wortman J, Feldgarden M, Lawley T, Gill SR, Haas BJ, Birren B, Gilmore MS. Kos VN, et al. mBio. 2012 May 22;3(3):e00112-12. doi: 10.1128/mBio.00112-12. Print 2012. mBio. 2012. PMID: 22617140 Free PMC article. - Methicillin-resistant Staphylococcus aureus: the European landscape.
Johnson AP. Johnson AP. J Antimicrob Chemother. 2011 May;66 Suppl 4:iv43-iv48. doi: 10.1093/jac/dkr076. J Antimicrob Chemother. 2011. PMID: 21521706 Review. - The evolution of Staphylococcus aureus.
Deurenberg RH, Stobberingh EE. Deurenberg RH, et al. Infect Genet Evol. 2008 Dec;8(6):747-63. doi: 10.1016/j.meegid.2008.07.007. Epub 2008 Jul 29. Infect Genet Evol. 2008. PMID: 18718557 Review.
Cited by
- Evolutionary Mechanisms of Long-Term Genome Diversification Associated With Niche Partitioning in Marine Picocyanobacteria.
Doré H, Farrant GK, Guyet U, Haguait J, Humily F, Ratin M, Pitt FD, Ostrowski M, Six C, Brillet-Guéguen L, Hoebeke M, Bisch A, Le Corguillé G, Corre E, Labadie K, Aury JM, Wincker P, Choi DH, Noh JH, Eveillard D, Scanlan DJ, Partensky F, Garczarek L. Doré H, et al. Front Microbiol. 2020 Sep 15;11:567431. doi: 10.3389/fmicb.2020.567431. eCollection 2020. Front Microbiol. 2020. PMID: 33042072 Free PMC article. - Molecular characterization of rifampicin-resistant Staphylococcus aureus isolates in a Chinese teaching hospital from Anhui, China.
Zhou W, Shan W, Ma X, Chang W, Zhou X, Lu H, Dai Y. Zhou W, et al. BMC Microbiol. 2012 Oct 22;12:240. doi: 10.1186/1471-2180-12-240. BMC Microbiol. 2012. PMID: 23082766 Free PMC article. - Explaining microbial phenotypes on a genomic scale: GWAS for microbes.
Dutilh BE, Backus L, Edwards RA, Wels M, Bayjanov JR, van Hijum SA. Dutilh BE, et al. Brief Funct Genomics. 2013 Jul;12(4):366-80. doi: 10.1093/bfgp/elt008. Epub 2013 Apr 26. Brief Funct Genomics. 2013. PMID: 23625995 Free PMC article. Review. - Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome.
Tomida S, Nguyen L, Chiu BH, Liu J, Sodergren E, Weinstock GM, Li H. Tomida S, et al. mBio. 2013 Apr 30;4(3):e00003-13. doi: 10.1128/mBio.00003-13. mBio. 2013. PMID: 23631911 Free PMC article. - Next generation deep sequencing and vaccine design: today and tomorrow.
Luciani F, Bull RA, Lloyd AR. Luciani F, et al. Trends Biotechnol. 2012 Sep;30(9):443-52. doi: 10.1016/j.tibtech.2012.05.005. Epub 2012 Jun 20. Trends Biotechnol. 2012. PMID: 22721705 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical