The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling - PubMed (original) (raw)
. 2010 Apr;84(7):3503-15.
doi: 10.1128/JVI.01161-09. Epub 2010 Jan 27.
Elena F Boer, Kirk J Lubick, James B Wolfinbarger, Aaron B Carmody, Barry Rockx, Wenjun Liu, Joseph Ashour, W Lesley Shupert, Michael R Holbrook, Alan D Barrett, Peter W Mason, Marshall E Bloom, Adolfo García-Sastre, Alexander A Khromykh, Sonja M Best
Affiliations
- PMID: 20106931
- PMCID: PMC2838099
- DOI: 10.1128/JVI.01161-09
The NS5 protein of the virulent West Nile virus NY99 strain is a potent antagonist of type I interferon-mediated JAK-STAT signaling
Maudry Laurent-Rolle et al. J Virol. 2010 Apr.
Abstract
Flaviviruses transmitted by arthropods represent a tremendous disease burden for humans, causing millions of infections annually. All vector-borne flaviviruses studied to date suppress host innate responses to infection by inhibiting alpha/beta interferon (IFN-alpha/beta)-mediated JAK-STAT signal transduction. The viral nonstructural protein NS5 of some flaviviruses functions as the major IFN antagonist, associated with inhibition of IFN-dependent STAT1 phosphorylation (pY-STAT1) or with STAT2 degradation. West Nile virus (WNV) infection prevents pY-STAT1 although a role for WNV NS5 in IFN antagonism has not been fully explored. Here, we report that NS5 from the virulent NY99 strain of WNV prevented pY-STAT1 accumulation, suppressed IFN-dependent gene expression, and rescued the growth of a highly IFN-sensitive virus (Newcastle disease virus) in the presence of IFN, suggesting that this protein can function as an efficient IFN antagonist. In contrast, NS5 from Kunjin virus (KUN), a naturally attenuated subtype of WNV, was a poor suppressor of pY-STAT1. Mutation of a single residue in KUN NS5 to the analogous residue in WNV-NY99 NS5 (S653F) rendered KUN NS5 an efficient inhibitor of pY-STAT1. Incorporation of this mutation into recombinant KUN resulted in 30-fold greater inhibition of JAK-STAT signaling than with the wild-type virus and enhanced KUN replication in the presence of IFN. Thus, a naturally occurring mutation is associated with the function of NS5 in IFN antagonism and may influence virulence of WNV field isolates.
Figures
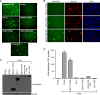
FIG. 1.
WNV NS5 antagonizes host IFN-β responses by inhibiting JAK-STAT signaling. (A) Effect of WNV NS5 expression on NDV-GFP replication in the presence of IFN. Vero cells transfected with an empty plasmid or a plasmid expressing individual virus proteins as indicated were treated with 1,000 U/ml IFN-β for 24 h and then infected with NDV-GFP. NDV-GFP replication was monitored by GFP fluorescence at 14 hpi. (B) Examples from panel A demonstrating NDV-GFP replication (green) in cells expressing flavivirus NS5 proteins (red). Nuclei are counter stained with 4′,6′-diamidino-2-phenylindole (DAPI; blue). (C) Western blot demonstrating the relative expression of each viral protein in HEK293T cells. (D) Effect of WNV NS5 on ISRE-dependent gene expression. HEK293T cells were transfected with the ISRE-CAT reporter plasmid, a plasmid constitutively expressing firefly luciferase, and either an empty plasmid or a plasmid expressing the virus protein indicated and treated with IFN for 24 h. CAT activity in cell lysates was determined and normalized to the luciferase activity in each sample. Fold induction of CAT activity was determined relative to the normalized CAT activity value of cells transfected with the empty vector and not treated with IFN. Error bars indicate standard errors of the mean (SEM) from three individual experiments performed in triplicate; asterisks indicate significant differences from cells transfected with the empty vector and not treated with IFN (P < 0.05, by ANOVA followed by Dunnett's multiple comparison test).
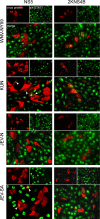
FIG. 2.
Immunofluorescence of STAT1 phosphorylation and nuclear localization in cells expressing flavivirus NS5 and 2KNS4B proteins. Vero cells were transfected with plasmids encoding the NS5 or 2KNS4B proteins derived from the indicated viruses, with each fused to a C-terminal V5 epitope tag. At 24 h posttransfection, cells were treated with IFN-β for 15 min, fixed in methanol, and stained for anti-V5 (red) and anti-pY-STAT1 (green). Arrowheads indicate examples of NS5-positive cells that are also positive for pY-STAT1.
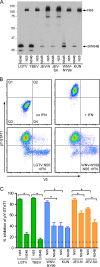
FIG. 3.
Quantification of inhibition of STAT1 phosphorylation by flavivirus NS5 and 2KNS4B proteins. (A) Western blot demonstrating the expression of NS5 and 2KNS4B proteins, each in frame with a C-terminal V5 epitope tag, in Vero cells. (B) Flow cytometry analysis to determine the percent inhibition of pY-STAT1 in Vero cells expressing flavivirus nonstructural proteins. Vero cells were fixed, permeabilized, and stained using antibodies that recognize pY(701)-STAT1 and V5. The examples shown are cells transfected with an empty plasmid and either untreated or treated with IFN-β (top panels) or cells expressing LGTV NS5 or WNV-NY99 NS5 and treated with IFN-β (lower panels). (C) Quantification of pY-STAT1 inhibition by each protein analyzed by flow cytometry. V5-positive cells were gated and examined for pY-STAT1. The percent pY-STAT1 inhibition is the percentage of V5-positive cells (contained in quadrant 2 [Q2] and Q4 in B) that were pY-STAT1 negative (in Q4). The average level of V5-negative cells that are also pY-STAT1 negative in IFN-β-treated cultures (cells present in Q3) is illustrated by the dotted line as an indication of the background levels in this assay. Error bars indicate SEM from four independent experiments; asterisks indicate significant differences (P < 0.05, by ANOVA followed by Tukey's test).
FIG. 4.
Identification of residues important for WNV NS5 function as an IFN antagonist. (A) Position of residues selected for mutation on the WNV KUN RdRp structure (31). The entire RdRp is shown on the left; the region of interest in IFN antagonism is shown on the right. The residues demonstrated in this study to have a role in WNV-NY99 NS5 function in IFN antagonism are depicted in red (W382, VI631/632, and W651) or green (residue 653), whereas those that were mutated to alanine with no effect on NS5 function are depicted in blue. (B) Western blot demonstrating the relative expression of each WNV-NY99 or KUN NS5 mutant used in these studies. (C) Quantification of pY-STAT1 inhibition by WNV-NY99 NS5 site mutants analyzed by flow cytometry. Error bars indicate SEM; asterisks indicate significant differences from WT NY99 NS5 (P < 0.05, by ANOVA followed by Dunnett's test). (D) IFA of WNV-NY99 NS5 containing a W651A mutation in IFN-β-treated Vero cells. NS5 is shown in red, and pY-STAT1 is shown in green.
FIG. 5.
Residue 653 is largely responsible for the differences between NY99 and KUN NS5 function in IFN antagonism. (A) Flow cytometry to determine the percent inhibition of pY-STAT1 in IFN-β-treated Vero cells expressing KUN and WNV-NY99 mutants. The examples shown are cells expressing WT KUN NS5, KUN NS5:S653F, WT WNV-NY99, and NY99 NS5:F653S. (B) Quantification of pY-STAT1 inhibition by WNV-NY99 and KUN NS5 mutants by flow cytometry. LGTV NS5 was included as a positive control for pY-STAT1 inhibition. Error bars indicate SEM from four independent experiments; asterisks indicate significant differences (P < 0.05, by ANOVA followed by Tukey's test).
FIG. 6.
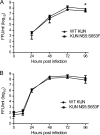
Replication of WT KUN and KUN NS5:S653F in cell culture. Supernatants from Vero cells infected at an MOI of 0.001 (A) or HEK293 cells infected at an MOI of 0.1 (B) were assayed for infectious virus by focus-forming assay at the times indicated. Error bars indicate SEM from three independent experiments performed in triplicate; the asterisk indicates significant difference (P < 0.05, Student's t test). FFU, focus-forming units.
FIG. 7.
Immunofluorescence of pY-STAT1 in WT and NS5:S653F KUN-infected cells. Vero cells were infected with WT or NS5:S653F KUN and treated with 1,000 U/ml IFN-β at 48 hpi, followed by staining for NS5 (red) and anti-pY-STAT1 (green). Nuclei were counterstained with DAPI.
FIG. 8.
Replication of NS5:S653F KUN is associated with greater IFN resistance than WT KUN. (A) Western blot demonstrating increased efficiency of IFN antagonism by KUN NS5:S653F. HEK293 cells were left uninfected or were infected with WT or NS5:S653F KUN at an MOI of 1. At the times indicated, cells were treated with 1,000 U/ml of IFN-β for 15 min, and cell lysates were analyzed for virus protein expression (NS5 and E) and STAT1 phosphorylation. (B) Effect of S653F mutation on ISRE-dependent gene expression during KUN replication. HEK293 cells were transfected with the pISRE-luc reporter plasmid and a plasmid constitutively expressing Renilla luciferase. Cells were infected 24 h later with WT or NS5:S653F KUN at an MOI of 1 and incubated for a further 24 h. Cells were treated with 1,000 U of IFN-β for 7 to 8 h prior to assaying for dual luciferase activities. Fold induction of firefly luciferase activity was determined relative to the normalized luciferase activity value of virus-infected cells not treated with IFN. Error bars indicate SEM from three individual experiments; asterisks indicate significant differences from cells transfected with the empty vector and not treated with IFN (P < 0.005, by Student's t test). (C) KUN replication in the presence of IFN. Vero cells were infected with WT or NS5:S653F KUN at an MOI of 0.001 and treated with high-dose IFN-β at 12 hpi. Supernatants were assayed for infectious virus by focus-forming assay at the times indicated. Error bars indicate SEM from five independent experiments performed in triplicate; an asterisk indicates significant difference (P = 0.0079, Mann-Whitney test). FFU, focus forming units.
Similar articles
- Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins.
Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. Liu WJ, et al. J Virol. 2005 Feb;79(3):1934-42. doi: 10.1128/JVI.79.3.1934-1942.2005. J Virol. 2005. PMID: 15650219 Free PMC article. - Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist.
Best SM, Morris KL, Shannon JG, Robertson SJ, Mitzel DN, Park GS, Boer E, Wolfinbarger JB, Bloom ME. Best SM, et al. J Virol. 2005 Oct;79(20):12828-39. doi: 10.1128/JVI.79.20.12828-12839.2005. J Virol. 2005. PMID: 16188985 Free PMC article. - A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice.
Liu WJ, Wang XJ, Clark DC, Lobigs M, Hall RA, Khromykh AA. Liu WJ, et al. J Virol. 2006 Mar;80(5):2396-404. doi: 10.1128/JVI.80.5.2396-2404.2006. J Virol. 2006. PMID: 16474146 Free PMC article. - The Many Faces of the Flavivirus NS5 Protein in Antagonism of Type I Interferon Signaling.
Best SM. Best SM. J Virol. 2017 Jan 18;91(3):e01970-16. doi: 10.1128/JVI.01970-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881649 Free PMC article. Review. - Suppression of Type I Interferon Signaling by Flavivirus NS5.
Thurmond S, Wang B, Song J, Hai R. Thurmond S, et al. Viruses. 2018 Dec 14;10(12):712. doi: 10.3390/v10120712. Viruses. 2018. PMID: 30558110 Free PMC article. Review.
Cited by
- The global ecology and epidemiology of West Nile virus.
Chancey C, Grinev A, Volkova E, Rios M. Chancey C, et al. Biomed Res Int. 2015;2015:376230. doi: 10.1155/2015/376230. Epub 2015 Mar 19. Biomed Res Int. 2015. PMID: 25866777 Free PMC article. Review. - Mosquito-borne flaviviruses and type I interferon: catch me if you can!
Zoladek J, Nisole S. Zoladek J, et al. Front Microbiol. 2023 Oct 30;14:1257024. doi: 10.3389/fmicb.2023.1257024. eCollection 2023. Front Microbiol. 2023. PMID: 37965539 Free PMC article. Review. - Type 1 IFN-independent activation of a subset of interferon stimulated genes in West Nile virus Eg101-infected mouse cells.
Pulit-Penaloza JA, Scherbik SV, Brinton MA. Pulit-Penaloza JA, et al. Virology. 2012 Apr 10;425(2):82-94. doi: 10.1016/j.virol.2012.01.006. Epub 2012 Feb 3. Virology. 2012. PMID: 22305622 Free PMC article. - Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling.
Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sánchez-Seco MP, Evans MJ, Best SM, García-Sastre A. Grant A, et al. Cell Host Microbe. 2016 Jun 8;19(6):882-90. doi: 10.1016/j.chom.2016.05.009. Epub 2016 May 19. Cell Host Microbe. 2016. PMID: 27212660 Free PMC article. - Current Understanding of West Nile Virus Clinical Manifestations, Immune Responses, Neuroinvasion, and Immunotherapeutic Implications.
Bai F, Thompson EA, Vig PJS, Leis AA. Bai F, et al. Pathogens. 2019 Oct 16;8(4):193. doi: 10.3390/pathogens8040193. Pathogens. 2019. PMID: 31623175 Free PMC article. Review.
References
- Arjona, A., M. Ledizet, K. Anthony, N. Bonafe, Y. Modis, T. Town, and E. Fikrig. 2007. West Nile virus envelope protein inhibits dsRNA-induced innate immune responses. J. Immunol. 179:8403-8409. - PubMed
- Beasley, D. W., M. C. Whiteman, S. Zhang, C. Y. Huang, B. S. Schneider, D. R. Smith, G. D. Gromowski, S. Higgs, R. M. Kinney, and A. D. Barrett. 2005. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J. Virol. 79:8339-8347. - PMC - PubMed
- Best, S. M., D. N. Mitzel, and M. E. Bloom. 2006. Action and reaction: the arthropod-borne flaviviruses and host interferon responses. Future Virol. 1:447-459.
Publication types
MeSH terms
Substances
Grants and funding
- F31 AI077333/AI/NIAID NIH HHS/United States
- FAI077333A/PHS HHS/United States
- U01 AI066321/AI/NIAID NIH HHS/United States
- 5UO1AI66321/AI/NIAID NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous