PHF6 mutations in T-cell acute lymphoblastic leukemia - PubMed (original) (raw)
doi: 10.1038/ng.542. Epub 2010 Mar 14.
Teresa Palomero, Hossein Khiabanian, Joni Van der Meulen, Mireia Castillo, Nadine Van Roy, Barbara De Moerloose, Jan Philippé, Sara González-García, María L Toribio, Tom Taghon, Linda Zuurbier, Barbara Cauwelier, Christine J Harrison, Claire Schwab, Markus Pisecker, Sabine Strehl, Anton W Langerak, Jozef Gecz, Edwin Sonneveld, Rob Pieters, Elisabeth Paietta, Jacob M Rowe, Peter H Wiernik, Yves Benoit, Jean Soulier, Bruce Poppe, Xiaopan Yao, Carlos Cordon-Cardo, Jules Meijerink, Raul Rabadan, Frank Speleman, Adolfo Ferrando
Affiliations
- PMID: 20228800
- PMCID: PMC2847364
- DOI: 10.1038/ng.542
PHF6 mutations in T-cell acute lymphoblastic leukemia
Pieter Van Vlierberghe et al. Nat Genet. 2010 Apr.
Abstract
Tumor suppressor genes on the X chromosome may skew the gender distribution of specific types of cancer. T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy with an increased incidence in males. In this study, we report the identification of inactivating mutations and deletions in the X-linked plant homeodomain finger 6 (PHF6) gene in 16% of pediatric and 38% of adult primary T-ALL samples. Notably, PHF6 mutations are almost exclusively found in T-ALL samples from male subjects. Mutational loss of PHF6 is importantly associated with leukemias driven by aberrant expression of the homeobox transcription factor oncogenes TLX1 and TLX3. Overall, these results identify PHF6 as a new X-linked tumor suppressor in T-ALL and point to a strong genetic interaction between PHF6 loss and aberrant expression of TLX transcription factors in the pathogenesis of this disease.
Figures
Figure 1
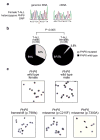
Next generation sequencing and array CGH analysis of the X chromosome identifies PHF6 mutations in human T-ALL. (a) Overview of mutation screening approach of the human X chromosome exome in a panel of tumor DNA samples from 12 male T-ALL cases using oligonucleotide sequence capture and next generation sequencing with SOLiD3. After filtering and confirmation of high throughput sequencing data, analysis of corresponding remission DNA samples led to the identification of three somatically acquired changes in the PHF6 gene. (b) Schematic overview of the recurrent genomic deletions involving chromosomal band Xq26.3 in 8 human T-ALL samples. Specific genes located in Xq26.3 are shown. (c) Detailed view of a representative oligo array-CGH plot of leukemia DNA/control DNA ratios (blue tracing) versus the dye-swap experiment (red tracing) in a patient harboring an Xq26.3 deletion. (d) DNA quantitative PCR analysis of PHF6 copy number dose in female and male reference genomic DNAs and 2 primary samples from male T-ALL cases harboring Xq26.3 deletions.
Figure 2
PHF6 mutations and expression in T-ALL lymphoblasts. (a) Structure of the PHF6 protein including four nuclear localization signals and two imperfect PHD zinc finger domains. Overview of all PHF6 mutations identified in primary T-ALL samples and T-ALL cell lines. Filled circles represent nonsense and frameshift mutations, whereas missense mutations are depicted as open circles. Circles filled in gray indicate mutations identified in female T-ALL cases. (b) Representative DNA sequencing chromatograms of paired diagnosis and remission genomic T-ALL DNA samples showing a somatic mutation in exon 7 of PHF6. (c) Western blot analysis of T-ALL cell lines revealed complete loss of PHF6 protein expression in the PHF6 mutated T-ALL cell lines. (d) PHF6 immunostaining in the Jurkat and HPB-ALL, wild-type and mutant T-ALL cell lines, respectively. (e) Western blot analysis of PHF6 and gamma-H2AX expression in HEK293T cells upon PHF6 shRNA knockdown. Actin levels are shown as loading control.
Figure 3
PHF6 expression in T-ALL lymphoblasts. (a) Sequence analysis of paired genomic DNA and cDNA samples shows monoallelic expression of PHF6 SNP rs17317724 in lymphoblasts from a wild-type PHF6 female T-ALL case. (b) Differential distribution of PHF6 mutations in T-ALL samples from male and female cases. (c) Immunohistochemical analysis of PHF6 expression in wild type and mutant T-ALL lymphoblasts.
Figure 4
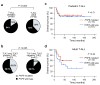
Clinical and biological characteristics associated with PHF6 mutations in T-ALL. (a) Frequencies of PHF6 mutations in pediatric and adult T-ALL samples. (b) Differential distribution of PHF6 mutations in TLX1/TLX3 positive and negative T-ALL samples. (c) Kaplan-Meier curve of overall survival in pediatric T-ALL patients from DCOG trials ALL7, ALL8 and ALL9 with and without PHF6 mutations. (d) Kaplan-Meier survival curve in adult T-ALL patients with and without mutations in PHF6 treated in ECOG clinical trial ECOG2993.
Similar articles
- Mutations of PHF6 are associated with mutations of NOTCH1, JAK1 and rearrangement of SET-NUP214 in T-cell acute lymphoblastic leukemia.
Wang Q, Qiu H, Jiang H, Wu L, Dong S, Pan J, Wang W, Ping N, Xia J, Sun A, Wu D, Xue Y, Drexler HG, Macleod RA, Chen S. Wang Q, et al. Haematologica. 2011 Dec;96(12):1808-14. doi: 10.3324/haematol.2011.043083. Epub 2011 Aug 31. Haematologica. 2011. PMID: 21880637 Free PMC article. Clinical Trial. - PHF6 mutations in adult acute myeloid leukemia.
Van Vlierberghe P, Patel J, Abdel-Wahab O, Lobry C, Hedvat CV, Balbin M, Nicolas C, Payer AR, Fernandez HF, Tallman MS, Paietta E, Melnick A, Vandenberghe P, Speleman F, Aifantis I, Cools J, Levine R, Ferrando A. Van Vlierberghe P, et al. Leukemia. 2011 Jan;25(1):130-4. doi: 10.1038/leu.2010.247. Epub 2010 Oct 29. Leukemia. 2011. PMID: 21030981 Free PMC article. - The Role of PHF6 in Hematopoiesis and Hematologic Malignancies.
Eisa YA, Guo Y, Yang FC. Eisa YA, et al. Stem Cell Rev Rep. 2023 Jan;19(1):67-75. doi: 10.1007/s12015-022-10447-4. Epub 2022 Aug 26. Stem Cell Rev Rep. 2023. PMID: 36008597 Review. - Structural and functional insights into the human Börjeson-Forssman-Lehmann syndrome-associated protein PHF6.
Liu Z, Li F, Ruan K, Zhang J, Mei Y, Wu J, Shi Y. Liu Z, et al. J Biol Chem. 2014 Apr 4;289(14):10069-83. doi: 10.1074/jbc.M113.535351. Epub 2014 Feb 19. J Biol Chem. 2014. PMID: 24554700 Free PMC article. - PHF6 Mutations in Hematologic Malignancies.
Kurzer JH, Weinberg OK. Kurzer JH, et al. Front Oncol. 2021 Jul 26;11:704471. doi: 10.3389/fonc.2021.704471. eCollection 2021. Front Oncol. 2021. PMID: 34381727 Free PMC article. Review.
Cited by
- LncRNA Bmp1 promotes the healing of intestinal mucosal lesions via the miR-128-3p/PHF6/PI3K/AKT pathway.
Zhuang M, Deng Y, Zhang W, Zhu B, Yan H, Lou J, Zhang P, Cui Q, Tang H, Sun H, Sun Y. Zhuang M, et al. Cell Death Dis. 2021 Jun 9;12(6):595. doi: 10.1038/s41419-021-03879-2. Cell Death Dis. 2021. PMID: 34108447 Free PMC article. - Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia.
Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, Sanchez-Martin M, Carpenter Z, Penson A, Perez-Garcia A, Eckert C, Nicolas C, Balbin M, Sulis ML, Kato M, Koh K, Paganin M, Basso G, Gastier-Foster JM, Devidas M, Loh ML, Kirschner-Schwabe R, Palomero T, Rabadan R, Ferrando AA. Oshima K, et al. Proc Natl Acad Sci U S A. 2016 Oct 4;113(40):11306-11311. doi: 10.1073/pnas.1608420113. Epub 2016 Sep 21. Proc Natl Acad Sci U S A. 2016. PMID: 27655895 Free PMC article. - PHF6 suppresses self-renewal of leukemic stem cells in AML.
Jalnapurkar SS, Pawar AS, George SS, Antony C, Somers P, Grana J, Feist VK, Gurbuxani S, Paralkar VR. Jalnapurkar SS, et al. Leukemia. 2024 Sep;38(9):1938-1948. doi: 10.1038/s41375-024-02340-5. Epub 2024 Jul 14. Leukemia. 2024. PMID: 39004675 Free PMC article. - Advances in the Diagnosis and Treatment of Pediatric Acute Lymphoblastic Leukemia.
Inaba H, Pui CH. Inaba H, et al. J Clin Med. 2021 Apr 29;10(9):1926. doi: 10.3390/jcm10091926. J Clin Med. 2021. PMID: 33946897 Free PMC article. Review. - Epigenetic Control of a Local Chromatin Landscape.
Chiarella AM, Lu D, Hathaway NA. Chiarella AM, et al. Int J Mol Sci. 2020 Jan 31;21(3):943. doi: 10.3390/ijms21030943. Int J Mol Sci. 2020. PMID: 32023873 Free PMC article. Review.
References
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. - PubMed
- Goldberg JM, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21:3616–3622. - PubMed
- Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. - PubMed
- Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54-AI057158/AI/NIAID NIH HHS/United States
- R01 CA129382/CA/NCI NIH HHS/United States
- R01CA120196/CA/NCI NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- R01CA129382/CA/NCI NIH HHS/United States
- R01 CA120196/CA/NCI NIH HHS/United States
- R01 CA129382-03/CA/NCI NIH HHS/United States
- 1R01LM010140-01/LM/NLM NIH HHS/United States
- U24 CA114737/CA/NCI NIH HHS/United States
- R01 LM010140/LM/NLM NIH HHS/United States
- R01 CA120196-03/CA/NCI NIH HHS/United States
- R01 CA155743/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials