Ajuba LIM proteins are negative regulators of the Hippo signaling pathway - PubMed (original) (raw)
Ajuba LIM proteins are negative regulators of the Hippo signaling pathway
Meghna Das Thakur et al. Curr Biol. 2010.
Abstract
The mammalian Ajuba LIM proteins (Ajuba, LIMD1, and WTIP) are adaptor proteins that exhibit the potential to communicate cell adhesive events with nuclear responses to remodel epithelia. Determining their role in vivo, however, has been challenging due to overlapping tissue expression and functional redundancy. Thus, we turned to Drosophila, where a single gene, CG11063 or djub, exists. Drosophila lacking the djub gene or depleted of dJub by RNA interference identify djub as an essential gene for development and a novel regulator of epithelial organ size as a component of the conserved Hippo (Hpo) pathway, which has been implicated in both tissue size control and cancer development. djub-deficient tissues were small and had decreased cell numbers as a result of increased apoptosis and decreased proliferation, due to downregulation of DIAP1 and cyclin E. This phenocopies tissues deficient for Yorkie (Yki), the downstream target of the Hippo pathway. djub genetically interacts with the Hippo pathway, and epistasis suggests that djub lies downstream of hpo. In mammalian and Drosophila cells, Ajuba LIM proteins/dJub interact with LATS/Warts (Wts) and WW45/Sav to inhibit phosphorylation of YAP/Yki. This work describes a novel role for the Ajuba LIM proteins as negative regulators of the Hippo signaling pathway.
Figures
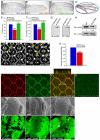
Figure 1. dJub regulates tissue size by controlling cell number
(A-C) Wings from wt female flies (A), female flies expressing dJub RNAi (B), or female flies expressing dJub-mCherry transgene (C). 1096-Gal4 was used to drive RNAi or transgene expression. (D) Outlines of the wings in panels A-C. (E, F) Quantification of relative wing areas (E) and cell numbers (F) of genotypes in A-C. Area and cell number measurements were taken from the wing region located between veins L4 and L5, and wt defined as 100% (N=20 for each). (G) Extracts of mammalian HEK293 cells transfected with myc-dJub immunoblotted with dJub antiserum (left) or Myc antiserum (right). (H) Immunoblot analysis of dJub protein levels in wt or dJub RNAi-expressing larval eye imaginal discs. Actin serves as loading control. Relative amount of dJub protein is indicated below each lane. Mid-pupal wt eyes (I) or GMR-Gal4 driven dJub RNAi expressing eyes (J) stained for DE-cadherin. Secondary (arrows) and tertiary (arrowheads) interommatidial cells are highlighted. Loss of interommatidial cells in dJub RNAi expressing pupal eyes denoted by arrows (J). (K) Quantification of relative numbers of interommatidial cells in wt versus dJub RNAi pupal eye. Interommatidial cells were counted in 20 fields, each containing a cluster of at least 7 ommatidia. (L-O) Mid-pupal wt eyes stained for DE-cadherin (L), dJub (M), and merged image (N). Z-stack analysis of line in N is shown above panel N (N). (O) Immunostaining with dJub antiserum preabsorbed with immunizing peptide. (P-R) Scanning electron micrographs (SEMs) of female adult eyes. WT (Q), _djub_I generated via the EGUF-Hid method, which results in eyes composed almost entirely of mutant tissue (Q), and GMR-gal4 driven overexpression of UAS-dJub-mCherry transgene (R). (S-U) Female third-instar larval eye imaginal discs containing wt (S) or _djub_I mutant (T, U) clones marked by the absence of GFP expression (black). (U) Enlarged view of _djub_I and wt twin spot clones. Yellow arrow identifies _djub_I clones, red arrow identifies wt twin spot clone containing two copies of Ubi-GFP, and white arrow identifies tissue carrying one copy of Ubi-GFP. In all experiments wings and eyes were dissected from female flies. In graphs data are shown as mean percentages +/− standard deviation, with N= 20 for each genotype. (***) Represents p-value ≤ 0.001 and (*) represents p-value ≤ 0.05. Anterior is to the left for all larval imaginal discs.
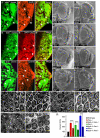
Figure 2. djub affects expression of DIAP1 and Cyclin E and genetically interacts with the Hippo pathway
(A-C) Female third-instar larval eye imaginal discs containing _djub_I clones (GFP –ve, yellow arrows) generated by Eyeless-flp and stained for DIAP1 (A’), DIAP1-lacZ (B’), or Cyclin E (C’) expression. Anterior is to the left for all larval eye imaginal discs. (D-K) SEMs of adult female Drosophila eyes of wt (E) and Hippo pathway mutants (F,H,J) and _djub_I (E) or Hippo pathway mutants containing a deletion of a single copy of djub (G,I,K), as indicated. Mid-pupal eye dissection of Hippo pathway mutants (L,N,P) or Hippo pathway mutants containing a deletion of a single copy of djub (M,O,Q), as indicated, and stained for DE-cadherin to identify interommatidial cells. Scale bars in (D-K) equal 100μm and (L-Q) equal 10μm. (R) Quantification of interommatidial cell numbers in 10 random fields containing 10 ommatidia each of genotypes (D,F-K). Data are shown as mean percentages +/− standard deviation and (***) represents p-value ≤ 0.001.
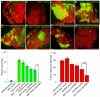
Figure 3. djub is epistatic to hpo based on clonal area and interommatidial cell numbers
(A-H) Female mid-pupal eyes stained for DE-cadherin (red). Wt, GFP positive MARCM clones (A); _djub_I MARCM clones (GFP +ve) (B); MARCM clones overexpressing Yki (GFP +ve) (C); MARCM clones mutant for _djub_I and overexpressing Yki (GFP +ve) (D); MARCM clones expressing wts RNAi (GFP +ve) (E); MARCM clones mutant for _djub_I and expressing wts RNAi (GFP +ve) (F); MARCM clones expressing hpo RNAi (GFP +ve) (G); MARCM clones mutant for _djub_I and expressing hpo RNAi (GFP +ve) (H). (I) Quantification of the clonal area (GFP +ve) for each genotype as a percentage of the entire pupal eye area. (J) Quantification of the percent increase of interommatidial cells within the clonal area (GFP +ve) as compared to wild type (set at 100% IOCs) for each genotype. In graphs data are shown as mean percentages +/− standard deviation, with N= 10 for each genotype. (***) Represents p-value ≤ 0.001. Scale bars in (A-H) equal 20μm.
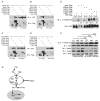
Figure 4. Ajuba LIM proteins associate with components of the Hippo pathway in mammalian cells and influence YAP phosphorylation
(A-D) HEK293 cells were co-transfected with LIMD1-YFP (A), Ajuba-YFP (B), Myc-WTIP (C), or Myc-Zyxin (D) and Flag-tagged Mst1, Lats1/2, WW45 or YAP, as indicated. Cell lysates were immunoprecipitated for each Hippo pathway member (anti-Flag), and bound products Immunoblotted (IB) for the presence of each LIM protein (anti-YFP or anti-Myc). Immunoblots of input controls (5%) are shown on the right side of each panel. (E) HEK293 cells were transfected with the indicated member of the Hippo pathway in the absence or presence of LIMD1-YFP. Levels of phospho-S127-YAP (upper panel) or total YAP (lower panels) were then determined by immunoblot (IB) analysis. Relative amounts of phospho-S127-YAP with respect to total YAP protein is indicated below each lane. (F) MDCK cells were transfected with control Luc siRNA (lanes 1-3) or Ajuba and LIMD1 siRNAs (lanes 4-6) and then plated at low (LD), medium (MD) and high density (HD). Amount of S127-YAP phosphorylation relative to total YAP for each density within control and Ajuba/LIMD1 depleted cells was determined by immunoblotting. The relative level of YAP phosphorylation for each density between control and Ajuba/LIMD1 depleted cells was determined and is indicated above the lanes. (G) Working model, based upon results herein, for how Ajuba LIM proteins could influence Hippo pathway signaling.
Similar articles
- Ajuba family proteins link JNK to Hippo signaling.
Sun G, Irvine KD. Sun G, et al. Sci Signal. 2013 Sep 10;6(292):ra81. doi: 10.1126/scisignal.2004324. Sci Signal. 2013. PMID: 24023255 Free PMC article. - The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway.
Wu S, Liu Y, Zheng Y, Dong J, Pan D. Wu S, et al. Dev Cell. 2008 Mar;14(3):388-98. doi: 10.1016/j.devcel.2008.01.007. Epub 2008 Feb 7. Dev Cell. 2008. PMID: 18258486 - The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control.
Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. Zhang L, et al. Dev Cell. 2008 Mar;14(3):377-87. doi: 10.1016/j.devcel.2008.01.006. Epub 2008 Feb 7. Dev Cell. 2008. PMID: 18258485 Free PMC article. - Hippo signaling pathway and organ size control.
Zhang L, Yue T, Jiang J. Zhang L, et al. Fly (Austin). 2009 Jan-Mar;3(1):68-73. doi: 10.4161/fly.3.1.7788. Epub 2009 Jan 7. Fly (Austin). 2009. PMID: 19164949 Free PMC article. Review. - A role for Hipk in the Hippo pathway.
Heidary Arash E, Attisano L. Heidary Arash E, et al. Sci Signal. 2013 May 14;6(275):pe18. doi: 10.1126/scisignal.2004259. Sci Signal. 2013. PMID: 23674821 Review.
Cited by
- YAP/TAZ enhances P-body formation to promote tumorigenesis.
Shen X, Peng X, Guo Y, Dai Z, Cui L, Yu W, Liu Y, Liu CY. Shen X, et al. Elife. 2024 Jul 24;12:RP88573. doi: 10.7554/eLife.88573. Elife. 2024. PMID: 39046443 Free PMC article. - Control of organ growth by patterning and hippo signaling in Drosophila.
Irvine KD, Harvey KF. Irvine KD, et al. Cold Spring Harb Perspect Biol. 2015 Jun 1;7(6):a019224. doi: 10.1101/cshperspect.a019224. Cold Spring Harb Perspect Biol. 2015. PMID: 26032720 Free PMC article. Review. - Hippo/YAP pathway for targeted therapy.
Felley-Bosco E, Stahel R. Felley-Bosco E, et al. Transl Lung Cancer Res. 2014 Apr;3(2):75-83. doi: 10.3978/j.issn.2218-6751.2014.02.03. Transl Lung Cancer Res. 2014. PMID: 25806284 Free PMC article. Review. - Identification and characterization of differentially expressed miRNAs in HepG2 cells under normoxic and hypoxic conditions.
Kong F, Ran W, Jiang N, Li S, Zhang D, Sun D. Kong F, et al. RSC Adv. 2019 May 29;9(29):16884-16891. doi: 10.1039/c9ra01523j. eCollection 2019 May 24. RSC Adv. 2019. PMID: 35516357 Free PMC article. - Support for the reproductive ground plan hypothesis of social evolution and major QTL for ovary traits of Africanized worker honey bees (Apis mellifera L.).
Graham AM, Munday MD, Kaftanoglu O, Page RE Jr, Amdam GV, Rueppell O. Graham AM, et al. BMC Evol Biol. 2011 Apr 13;11:95. doi: 10.1186/1471-2148-11-95. BMC Evol Biol. 2011. PMID: 21489230 Free PMC article.
References
- Marie H, Pratt SJ, Betson M, Epple H, Kittler JT, Meek L, Moss SJ, Troyanovsky S, Attwell D, Longmore GD, et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem. 2003;278:1220–1228. - PubMed
- Iida S, Hirota T, Morisaki T, Marumoto T, Hara T, Kuninaka S, Honda S, Kosai K, Kawasuji M, Pallas DC, et al. Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene. 2004;23:5266–5274. - PubMed
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. - PubMed
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM080673-03/GM/NIGMS NIH HHS/United States
- R01 NS036570/NS/NINDS NIH HHS/United States
- GM080673/GM/NIGMS NIH HHS/United States
- R01 GM080673/GM/NIGMS NIH HHS/United States
- R01 CA085839-09/CA/NCI NIH HHS/United States
- CA85839/CA/NCI NIH HHS/United States
- R01 GM068048/GM/NIGMS NIH HHS/United States
- GM068048/GM/NIGMS NIH HHS/United States
- R01 CA085839/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases