Comparative metagenomics and population dynamics of the gut microbiota in mother and infant - PubMed (original) (raw)
Comparative metagenomics and population dynamics of the gut microbiota in mother and infant
Parag A Vaishampayan et al. Genome Biol Evol. 2010.
Abstract
Colonization of the gastrointestinal tract (GIT) of human infants with a suitable microbial community is essential for numerous aspects of health, but the progression of events by which this microbiota becomes established is poorly understood. Here, we investigate two previously unexplored areas of microbiota development in infants: the deployment of functional capabilities at the community level and the population genetics of its most abundant genera. To assess the progression of the infant microbiota toward an adult-like state and to evaluate the contribution of maternal GIT bacteria to the infant gut, we compare the infant's microbiota with that of the mother at 1 and 11 months after delivery. These comparisons reveal that the infant's microbiota rapidly acquires and maintains the range of gene functions present in the mother, without replicating the phylogenetic composition of her microbiota. Microdiversity analyses for Bacteroides and Bifidobacterium, two of the main microbiota constituents, reveal that by 11 months, the phylotypes detected in the infant are distinct from those in the mother, although the maternal Bacteroides phylotypes were transiently present at 1 month of age. The configuration of genetic variants within these genera reveals populations far from equilibrium and likely to be undergoing rapid growth, consistent with recent population turnovers. Such compositional turnovers and the associated loss of maternal phylotypes should limit the potential for long-term coadaptation between specific bacterial and host genotypes.
Keywords: Bacteroides; Bifidobacterium; bacterial population genetics; community genomics; gut microbiota.
Figures
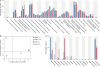
FIG. 1.—
Phylogenetic composition and general functional capabilities across GIT microbiota samples. (A) Distribution of COG functional categories in the GIT microbiota of mother and infant at 1 and 11 months after delivery. Distributions of COG functional classes are remarkably similar; observed differences across samples do not reach significance (D rank analysis; P > 0.05). (B) PCA of overall COG category profiles in the mother–infant GIT microbiota. Two additional samples (black and gray circles) from adult humans (Gill et al. 2006) are included for comparative purposes. Note the overlapping positions of the two maternal microbiota samples and the infant sample taken at 11 months. The large separation of I-1m from these three samples on the second PC axis stems from the fact that this is the only sample containing the COG category Extracellular Structures (W); the first PC axis separates the four samples in our study from those of Gill et al. (2006) due to the higher numbers of individual COGs represented within each category in these larger samples. (C) Bacterial diversity present in the gastrointestinal microbiota of mother and infant at 1 and 11 months after delivery. Taxonomic affiliation was assigned to the high-scoring (>90% identity) best BlastP matches of protein-coding genes in fosmid-end sequence reads against IMG/M reference isolate genomes. Most identified genera belong to only four phyla: Bacteroidetes (Bacteroides), Actinobacteria (Bifidobacterium, Collinsella, and_Eggerthella_), Proteobacteria (Escherichia, Klebsiella,Shigella, Salmonella,Citrobacter, and other enterics), and Firmicutes (Clostridium, Dorea, other clostridia, and Eubacterium).
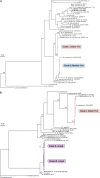
FIG. 2.—
Phylogeny of GIT haplotypes in relation to diversity within the corresponding genus. Phylogenies of the (A)Bifidobacterium and (B)Bacteroides 16S rRNA haplotypes with representative samples from each genus were derived by maximum likelihood with 1,000 bootstrap replicates under a general time-reversible model with invariant sites and gamma-distributed site rate variation. Scale bar represents 0.02 nucleotide substitutions per site. “Mixed” clades contain haplotypes recovered from both the mother and the infant.
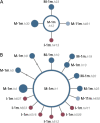
FIG. 3.—
Genealogy of Bacteroides haplotypes. (A) Haplotypes forming clade II in figure 2_B_, including haplotypes isolated from the mother at 1 month after delivery (M-1m; dark blue circles) as well as new haplotypes not present in the M-1m sample but detected in the infant at 1 month (I-1m; red) or the mother at 11 months after delivery (M-11m; light blue). (B) Haplotypes forming clade III in figure 2_B_, following the same color scheme described above. Haplotypes are numbered with the prefix “h.” Area of circles denotes haplotype frequency in the M-1m, M-11m, and I-1m samples combined; the smallest circles represent singletons and the large central circles the likely ancestors from which other haplotypes derive. Unmodified lines connecting haplotypes indicate single mutational steps, and tick marks on the lines indicate additional steps. Note that most derived haplotypes differ from the ancestor by a single mutational step.
Similar articles
- Metagenomics and development of the gut microbiota in infants.
Vallès Y, Gosalbes MJ, de Vries LE, Abellán JJ, Francino MP. Vallès Y, et al. Clin Microbiol Infect. 2012 Jul;18 Suppl 4:21-6. doi: 10.1111/j.1469-0691.2012.03876.x. Clin Microbiol Infect. 2012. PMID: 22647043 - Impact of early events and lifestyle on the gut microbiota and metabolic phenotypes in young school-age children.
Zhong H, Penders J, Shi Z, Ren H, Cai K, Fang C, Ding Q, Thijs C, Blaak EE, Stehouwer CDA, Xu X, Yang H, Wang J, Wang J, Jonkers DMAE, Masclee AAM, Brix S, Li J, Arts ICW, Kristiansen K. Zhong H, et al. Microbiome. 2019 Jan 4;7(1):2. doi: 10.1186/s40168-018-0608-z. Microbiome. 2019. PMID: 30609941 Free PMC article. - The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review.
Rutayisire E, Huang K, Liu Y, Tao F. Rutayisire E, et al. BMC Gastroenterol. 2016 Jul 30;16(1):86. doi: 10.1186/s12876-016-0498-0. BMC Gastroenterol. 2016. PMID: 27475754 Free PMC article. Review. - Influence of mother's intestinal microbiota on gut colonization in the infant.
Grönlund MM, Grześkowiak Ł, Isolauri E, Salminen S. Grönlund MM, et al. Gut Microbes. 2011 Jul-Aug;2(4):227-33. doi: 10.4161/gmic.2.4.16799. Epub 2011 Jul 1. Gut Microbes. 2011. PMID: 21983067 Clinical Trial. - Opportunities for improving the health and nutrition of the human infant by probiotics.
Salminen S, Isolauri E. Salminen S, et al. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:223-33; discussion 233-7. doi: 10.1159/000146350. Nestle Nutr Workshop Ser Pediatr Program. 2008. PMID: 18626203 Review.
Cited by
- Transcriptional and functional characterization of genetic elements involved in galacto-oligosaccharide utilization by Bifidobacterium breve UCC2003.
O'Connell Motherway M, Kinsella M, Fitzgerald GF, van Sinderen D. O'Connell Motherway M, et al. Microb Biotechnol. 2013 Jan;6(1):67-79. doi: 10.1111/1751-7915.12011. Epub 2012 Dec 2. Microb Biotechnol. 2013. PMID: 23199239 Free PMC article. - Genetic improvement and genomic resources of important cyprinid species: status and future perspectives for sustainable production.
Rasal KD, Kumar PV, Risha S, Asgolkar P, Harshavarthini M, Acharya A, Shinde S, Dhere S, Rasal A, Sonwane A, Brahmane M, Sundaray JK, Nagpure N. Rasal KD, et al. Front Genet. 2024 Sep 19;15:1398084. doi: 10.3389/fgene.2024.1398084. eCollection 2024. Front Genet. 2024. PMID: 39364006 Free PMC article. Review. - Microbial ecology and host-microbiota interactions during early life stages.
Collado MC, Cernada M, Baüerl C, Vento M, Pérez-Martínez G. Collado MC, et al. Gut Microbes. 2012 Jul-Aug;3(4):352-65. doi: 10.4161/gmic.21215. Epub 2012 Jun 29. Gut Microbes. 2012. PMID: 22743759 Free PMC article. Review. - Microbial embryonal colonization during pipefish male pregnancy.
Beemelmanns A, Poirier M, Bayer T, Kuenzel S, Roth O. Beemelmanns A, et al. Sci Rep. 2019 Jan 9;9(1):3. doi: 10.1038/s41598-018-37026-3. Sci Rep. 2019. PMID: 30626884 Free PMC article. - Human behavior, not race or geography, is the strongest predictor of microbial succession in the gut bacteriome of infants.
Quin C, Gibson DL. Quin C, et al. Gut Microbes. 2020 Sep 2;11(5):1143-1171. doi: 10.1080/19490976.2020.1736973. Epub 2020 Apr 5. Gut Microbes. 2020. PMID: 32249675 Free PMC article. Review.
References
- Adlerberth I, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. 2006;59:96–101. - PubMed
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases