53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks - PubMed (original) (raw)
Comment
. 2010 Apr 16;141(2):243-54.
doi: 10.1016/j.cell.2010.03.012. Epub 2010 Apr 1.
Elsa Callén, Nancy Wong, Hua-Tang Chen, Federica Polato, Amanda Gunn, Anne Bothmer, Niklas Feldhahn, Oscar Fernandez-Capetillo, Liu Cao, Xiaoling Xu, Chu-Xia Deng, Toren Finkel, Michel Nussenzweig, Jeremy M Stark, André Nussenzweig
Affiliations
- PMID: 20362325
- PMCID: PMC2857570
- DOI: 10.1016/j.cell.2010.03.012
Comment
53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks
Samuel F Bunting et al. Cell. 2010.
Abstract
Defective DNA repair by homologous recombination (HR) is thought to be a major contributor to tumorigenesis in individuals carrying Brca1 mutations. Here, we show that DNA breaks in Brca1-deficient cells are aberrantly joined into complex chromosome rearrangements by a process dependent on the nonhomologous end-joining (NHEJ) factors 53BP1 and DNA ligase 4. Loss of 53BP1 alleviates hypersensitivity of Brca1 mutant cells to PARP inhibition and restores error-free repair by HR. Mechanistically, 53BP1 deletion promotes ATM-dependent processing of broken DNA ends to produce recombinogenic single-stranded DNA competent for HR. In contrast, Lig4 deficiency does not rescue the HR defect in Brca1 mutant cells but prevents the joining of chromatid breaks into chromosome rearrangements. Our results illustrate that HR and NHEJ compete to process DNA breaks that arise during DNA replication and that shifting the balance between these pathways can be exploited to selectively protect or kill cells harboring Brca1 mutations.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
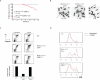
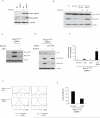
Figure 1. Deletion of 53BP1 reduces mammary tumorigenesis, radial chromosome formation and cellular proliferation defects in Brca1Δ11/Δ11 cells
(A) Breast cancer incidence in mice deficient in Brca1. The red line indicates tumor incidence in mice with deletion of Brca1 exon 11 targeted to the mammary cells using a Cre transgene under control of the MMTV LTR promoter (Brca1ϕ/ϕ;MMTV-Cre). The blue line shows mammary tumor incidence in Brca1 Δ11/Δ11 _53BP1_-/- mice, in which Brca1 exon 11 and 53BP1 are deleted in all cells. (B) Radial chromosome structures characteristic of metaphases from Brca1 Δ11/Δ11 p53+/- B cells. The percentage of metaphases containing radial chromosomes in Brca1 Δ11/Δ11 p53+/- (n=300), Brca1 Δ11/Δ11 _53BP1_-/- (n=300) and WT (n=100) cells is indicated. Note that equivalent results were seen with Brca1 Δ11/Δ11 p53+/- and Brca1 Δ11/Δ11 _p53_-/-cells. (C) Flow cytometry analysis of B cells to measure cell survival. The labeled populations in the dot plots are viable cells, identified by their ability to exclude PI and their lack of caspase 3 activation. The chart shows the frequency of live cells treated with PARP inhibitor normalized to the untreated population. (D) Proliferation of B cells pulsed with CFSE and cultured with and without PARP inhibitor. CFSE signal diminishes with increasing cell division, so that cells that do not divide have the highest CFSE signal. See also Fig. S1.
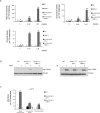
Figure 2. Deletion of 53BP1 reverses sensitivity of Brca1Δ11/Δ11 cells to PARPi and camptothecin
(A) Analysis of genomic instability in metaphases from B cells treated with 0, 10 nM and 1μM PARP inhibitor. Charts show the number of radial chromosomes, chromatid breaks and chromosome breaks per 100 metaphases (n=50 metaphases analyzed in each case). Note that genomic instability in Brca1 Δ11/Δ11 cells is independent of p53 status and equivalent results were seen in Brca1 Δ11/Δ11 p53+/- and Brca1 Δ11/Δ11 _p53_-/- cells. (B) Western blot showing Kap1 phosphorylation in B cells from the indicated genotypes treated with PARP inhibitor or 5Gy ionizing radiation. (C) Analysis of genomic instability in metaphases from B cells treated with 4nM camptothecin (CPT). B cells were cultured overnight with CPT prior to fixation and preparation of metaphase slides.
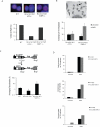
Figure 3. Deletion of 53BP1 restores HR in Brca1- but not _Xrcc2_- mutant cells
(A) Immunofluorescence images showing Rad51 foci (red) with DAPI counterstain (blue) in cells of the indicated genotypes after treatment with ionizing radiation. Chart shows the percentage of cells with Rad51 foci (n=100 counted for each genotype). (B) B cells grown for 36 hours in BrdUTP were fixed and metaphases prepared to visualize individual sister chromatids. Sister chromatid exchanges (SCEs) in metaphase chromosomes are indicated with arrows in the image in the top panel. The chart shows the number of SCEs in B cells of the indicated genotypes treated with and without PARP inhibitor. (C) Upper panel: Structure of the HR reporter substrate DR-GFPhyg is shown, along with the HR product expressing a functional GFP+ gene. Lower panel: Frequency of HR relative to total transfected cells in WT, Brca1 _Δ11/Δ_11 and Brca1 Δ11/Δ11 _53BP1_-/- MEFs, as measured using the DR-GFPhyg reporter. *p<0.0001 between WT and Brca1 Δ11/Δ11, *p<0.0028 between WT and Brca1 Δ11/Δ11 _53BP1_-/-. (D) Analysis of genomic instability in metaphases from B cells treated with and without PARP inhibitor (1μM). B cells were homozygous for a conditional Xrcc2 allele that was deleted using a B cell-specific CD19-Cre transgene to produce knockout Xrcc2 ϕ/ϕ, and Xrcc2 ϕ/ϕ _53BP1_-/- cells. Charts show the number of radial chromosomes, chromatid breaks and chromosome breaks per 100 metaphases (n=50 metaphases analyzed in each case). See also Fig S2.
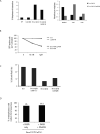
Figure 4. Deletion of Lig4 inhibits formation of radial chromosomes but does not rescue cell proliferation or genomic instability in Brca1 Δ11/Δ11 cells
(A) Analysis of genomic instability in metaphases from B cells treated with PARP inhibitor (1μM). Conditional Brca1 and Lig4 alleles were deleted using pMX-Cre-GFP retroviral transduction to generate B cells that were homozygous for the knockout Brca1 ϕ/ϕ and Lig4 ϕ/ϕ alleles. Chart (left) shows percentage of metaphases with genomic instability from cells of the indicated genotypes. Chart (right) quantifies the distribution of radial chromosomes, chromatid breaks (ctd) and chromosome breaks (csb) among the aberrant metaphases (n=50 metaphases of each genotype analyzed). (B) Cell survival after treatment with PARP inhibitor. Cultured B cells homozygous for conditional Brca1 and Lig4 alleles were infected with pMX-Cre-GFP retrovirus to produce GFP+ knockout Brca1 ϕ/ϕ and Lig4 ϕ/ϕ cells. The chart shows the percentage of GFP+ cells that were present in B cell cultures of the indicated genotypes five days post-infection. PARP inhibitor treatment (10nM or 1μM) led to disappearance of Brca1 ϕ/ϕ and Brca1 ϕ/ϕ Lig4 ϕ/ϕ cells from the cultures. (C) Chart showing the percentage of cells with Rad51 foci after ionizing radiation as measured by immunofluorescence (n ≥ 160 cells). (D) Frequency of radial chromosomes in metaphase spreads from Brca1 Δ11/Δ11 p53+/- B cells cultured for 24 hours with either PARP inhibitor (PARPi) or PARPi + DNA-PKcs inhibitor (DNAPKi).
Figure 5. Rescue of homologous recombination in Brca1 Δ11/Δ11 cells by 53BP1 deletion correlates with increased phosphorylation of RPA and is dependent on ATM and CtIP
(A) Western blot analysis showing Kap1 and RPA phosphorylation in WT and _53BP1_-/- cells in response to 30Gy ionizing radiation. (B) RPA and Kap1 phosphorylation after 30Gy ionizing radiation in _Brca1_- and _Lig4_-deficient cells. Knockout Brca1 ϕ/ϕ and Lig4 ϕ/ϕ cells were prepared by sorting B cells homozygous for the conditional alleles after infection with pMX-Cre-GFP retrovirus. (C) Western blot showing Kap1 and RPA phosphorylation in Brca1 Δ11/Δ11 _53BP1_-/- cells after 30Gy ionizing radiation with and without ATM inhibitor (KU55993). ATMi (5 μM) was added 2 hours prior to irradiation. (D) Western blot analysis showing ionizing radiation-induced RPA phosphorylation in Brca1 Δ11/Δ11 _53BP1_-/- MEFs after CtIP shRNA. Protein lysates were prepared from B cells infected with either vector (-) or CtIP shRNA (CTIP), with and without 30 Gy ionizing radiation. (E) Quantification of radial chromosome structures in metaphases from Brca1 Δ11/Δ11 _53BP1_-/- cells treated with ATM inhibitor (5 μM) and / or PARP inhibitor (1 μM). ATMi was added at the start of the B cell culture, PARPi at 24 hours, and metaphases were made at 48 hours (n ≥ 240 metaphases; mean +/- s.d. shown from two experiments). (F) CFSE dilution analysis showing proliferation of B cells cultured with ATM inhibitor and/or PARP inhibitor for 96 hours. (G) Quantification of Rad51 foci in Brca1 Δ11/Δ11 _53BP1_-/- cells. Rad51 foci were induced with 5Gy of ionizing radiation and cells fixed for Rad51 staining 2 hours later. Cells were either untreated, or pre-treated for 2hrs with 5μM ATM inhibitor. See also Fig. S3.
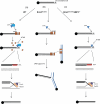
Figure 6. Model for reversal of genomic instability in Brca1 Δ11/Δ11 cells by 53BP1 deletion
Chromatid breaks accumulate in S-phase cells as a consequence of errors in DNA replication or through failure of single-strand break repair (e.g. after PARP inhibition). (A) In WT cells, Brca1 displaces 53BP1 from DSBs, enabling resection at the break site by factors such as CtIP, which promotes RPA loading onto single-stranded regions of DNA. RPA is displaced by Rad51, which enables strand invasion at the homologous region of the sister chromatid. Rad51 acts in complex with several effectors of HR, including Xrcc2, leading to error-free, template-directed repair of the double-strand break. In Brca1 Δ11/Δ11 cells, 53BP1 is not displaced, and inhibits resection. In the absence of resection, the break persists and if more than one chromatid break is present, these breaks can be joined by a Lig4-dependent NHEJ pathway. Joining of the chromatid breaks produces radial chromosome structures. (C) In Brca1 Δ11/Δ11 _53BP1_-/- cells, 53BP1 is not present at the double-strand break site, enabling resection even in Brca1-deficient cells. RPA and downstream effectors of homologous recombination are able to load normally at the break site, permitting error-free DNA repair. See also Fig. S4.
Comment on
- Loss of 53BP1 is a gain for BRCA1 mutant cells.
Kass EM, Moynahan ME, Jasin M. Kass EM, et al. Cancer Cell. 2010 May 18;17(5):423-5. doi: 10.1016/j.ccr.2010.04.021. Cancer Cell. 2010. PMID: 20478525
Similar articles
- ZMYM2 restricts 53BP1 at DNA double-strand breaks to favor BRCA1 loading and homologous recombination.
Lee D, Apelt K, Lee SO, Chan HR, Luijsterburg MS, Leung JWC, Miller KM. Lee D, et al. Nucleic Acids Res. 2022 Apr 22;50(7):3922-3943. doi: 10.1093/nar/gkac160. Nucleic Acids Res. 2022. PMID: 35253893 Free PMC article. - Ectopic expression of RNF168 and 53BP1 increases mutagenic but not physiological non-homologous end joining.
Zong D, Callén E, Pegoraro G, Lukas C, Lukas J, Nussenzweig A. Zong D, et al. Nucleic Acids Res. 2015 May 26;43(10):4950-61. doi: 10.1093/nar/gkv336. Epub 2015 Apr 27. Nucleic Acids Res. 2015. PMID: 25916843 Free PMC article. - CtIP-mediated resection is essential for viability and can operate independently of BRCA1.
Polato F, Callen E, Wong N, Faryabi R, Bunting S, Chen HT, Kozak M, Kruhlak MJ, Reczek CR, Lee WH, Ludwig T, Baer R, Feigenbaum L, Jackson S, Nussenzweig A. Polato F, et al. J Exp Med. 2014 Jun 2;211(6):1027-36. doi: 10.1084/jem.20131939. Epub 2014 May 19. J Exp Med. 2014. PMID: 24842372 Free PMC article. - Roles for the DNA-PK complex and 53BP1 in protecting ends from resection during DNA double-strand break repair.
Shibata A, Jeggo PA. Shibata A, et al. J Radiat Res. 2020 Sep 8;61(5):718-726. doi: 10.1093/jrr/rraa053. J Radiat Res. 2020. PMID: 32779701 Free PMC article. Review. - BRCA1, PARP, and 53BP1: conditional synthetic lethality and synthetic viability.
Aly A, Ganesan S. Aly A, et al. J Mol Cell Biol. 2011 Feb;3(1):66-74. doi: 10.1093/jmcb/mjq055. J Mol Cell Biol. 2011. PMID: 21278454 Free PMC article. Review.
Cited by
- Analysis for type of 53BP1 nuclear expression by immunofluorescence as an indicator of genomic instability in oropharyngeal squamous epithelial lesions.
Nishi H, Matsuda K, Terakado M, Kondo H, Kumai Y, Nakashima M. Nishi H, et al. Sci Rep. 2024 Nov 11;14(1):27525. doi: 10.1038/s41598-024-77945-y. Sci Rep. 2024. PMID: 39528680 Free PMC article. - PARP inhibitors in prostate cancer: clinical applications.
Saeidi H, Sarafbidabad M. Saeidi H, et al. Mol Biol Rep. 2024 Oct 30;51(1):1103. doi: 10.1007/s11033-024-10034-5. Mol Biol Rep. 2024. PMID: 39476131 Review. - Causes and consequences of DNA double-stranded breaks in cardiovascular disease.
Marian AJ. Marian AJ. Mol Cell Biochem. 2024 Oct 15. doi: 10.1007/s11010-024-05131-9. Online ahead of print. Mol Cell Biochem. 2024. PMID: 39404936 Review. - 53BP1 deficiency leads to hyperrecombination using break-induced replication (BIR).
Shah SB, Li Y, Li S, Hu Q, Wu T, Shi Y, Nguyen T, Ive I, Shi L, Wang H, Wu X. Shah SB, et al. Nat Commun. 2024 Oct 5;15(1):8648. doi: 10.1038/s41467-024-52916-z. Nat Commun. 2024. PMID: 39368985 Free PMC article. - NEAT1 modulates the TIRR/53BP1 complex to maintain genome integrity.
Kilgas S, Syed A, Toolan-Kerr P, Swift ML, Roychoudhury S, Sarkar A, Wilkins S, Quigley M, Poetsch AR, Botuyan MV, Cui G, Mer G, Ule J, Drané P, Chowdhury D. Kilgas S, et al. Nat Commun. 2024 Sep 30;15(1):8438. doi: 10.1038/s41467-024-52862-w. Nat Commun. 2024. PMID: 39349456 Free PMC article.
References
- Andreassen PR, Ho GP, D'Andrea AD. DNA damage responses and their many interactions with the replication fork. Carcinogenesis. 2006;27:883–892. - PubMed
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. - PMC - PubMed
- Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. - PubMed
- Brodie SG, Xu X, Qiao W, Li WM, Cao L, Deng CX. Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene. 2001;20:7514–7523. - PubMed
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI037526/AI/NIAID NIH HHS/United States
- R01 CA120954-04/CA/NCI NIH HHS/United States
- Z01 BC010283-10/ImNIH/Intramural NIH HHS/United States
- R01 CA120954/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- Z99 CA999999/ImNIH/Intramural NIH HHS/United States
- Z01 BC010959-01/ImNIH/Intramural NIH HHS/United States
- R01CA120954/CA/NCI NIH HHS/United States
- AI037526/AI/NIAID NIH HHS/United States
- R37 AI037526/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous