A tale of two pili: assembly and function of pili in bacteria - PubMed (original) (raw)
Review
A tale of two pili: assembly and function of pili in bacteria
Kimberly A Kline et al. Trends Microbiol. 2010 May.
Abstract
Bacterial pili have long been recognized as mediators of initial host-pathogen interactions important for the progression of Gram-negative bacterial diseases. An appreciation of the role of pili on virulence in Gram-positive bacteria and the unique properties of their biogenesis is a rapidly emerging area of research. In this review, we focus on recent advances in one of the longest-studied Gram-negative pilus systems, the chaperone/usher assembled pili, along with the newcomer to the field, the sortase-assembled pili of Gram-positive bacteria. In both systems, a wealth of new structural and molecular details has emerged recently. In light of this, we explore similarities between chaperone/usher and sortase-assembled pilus biogenesis and highlight paradigms unique to each, with the goal of using knowledge of each system to raise new questions and inform future studies of the other.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
Figure 1. CU and SA pilus and fiber assembly components
(a)–(b) E. coli prototype CU assembled pili. (a) Pilus subunit location within wild-type pilus fibers are depicted, as determined by immunoelectron microscopy. “*” indicates subunits whose location was deduced from biochemical and/or structural studies and not by immunoelectron microscopy. Empty boxes indicate no available data for that pilus biogenesis component. (b) Schematic of pilus biogenesis machinery for CU assembled pili. E. coli CU pilin subunits are translocated via the Sec machinery through the inner membrane (i), after which they associate with dedicated chaperone proteins in the periplasm (ii) which prevent subunit misfolding and facilitate delivery of the subunits to the outer membrane usher protein (iii) through which subunits are secreted and which serves as a platform for ordered pilus assembly (iv). (c)–(d) Gram-positive SA pili. (c) Table of pilus subunits for Gram-positive SA pili. Subscript for S. pyogenes indicates M1, M3 or M6 serotype and for S. agalactiae indicates the PI-1 or PI-2a pilus island. Slash marks (/) indicate differing names for the same protein in the literature. (d) Schematic of Gram-positive SA pili. Gram-positive pilin subunits are targeted for translocation across the cell membrane by the general secretion machinery via an N-terminal signal sequence (i). Presumably, subunits are then retained in the cell membrane by a transmembrane domain (ii) prior to LPXTG recognition and cleavage by sortase enzymes leading to ordered pilus fiber assembly (iii) and attachment to the cell wall (iv).
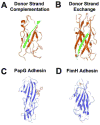
Figure 2. CU pilin subunit structure
(a) Donor strand complementation. The interactive subunit assembly site created by the incomplete immunoglobulin fold of the PapE (orange) pilin subunit is capped by the G1 strand of the PapD chaperone (green), providing the steric information necessary to complement the Ig-like fold of the subunit and stabilize the subunit prior to pilus assembly (PDB code 1NOL). (b) Donor strand exchange. PapE (orange) pilin structure consists of an incomplete Ig fold, which is completed within the pilus structure by the N-terminal extension of the neighboring pilin subunit, PapK (green), to form a canonical Ig domain in the polymerized pilus fiber (PDB code 1N12). (c) PapG ligand-binding domain (blue) bound to GbO4 (red), which consists of the PapG ligand tetrasaccharide GalNAc beta 1–3Gal alpha 1–4Gal beta 1–4Glc linked to ceramide (PDB code 1J8R). (d) FimH ligand-binding domain interacting with its D-mannose ligand (red) (PDB code 1KLF).
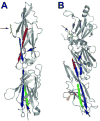
Figure 3. SA pilin subunit structure
(a) The 32.5 kD two-domain S. pyogenes major pilin subunit structure in ribbon diagram (PDB code 3B2m). The N- and C-terminal β sheets of the molecule are colored red and green, respectively. The interacting β sheet for each amide bond is colored blue. Stick diagrams indicate Lys-Asn intramolecular bonds within each domain of the subunit (indicated by large black arrows) and the conserved lysine (yellow, small black arrow) that becomes covalently linked to the threonine of cleaved LPXTG-like motif in the neighboring subunit. (b) The 47 kD three-domain C. diphtheriae SpaA major pilin subunit structure in ribbon diagram (PDB code 3HR6). The N- and C-terminal β sheets that participate in intramolecular amide bonds are colored red and green (in the middle and C-terminal domain, respectively); the interacting β sheet for each amide bond is colored blue. Stick diagrams indicate Lys-Asn intramolecular bonds within each domain of the subunit (large black arrows), the conserved lysine of the YPKN pilin motif (yellow, small black arrow) that becomes covalently linked to the threonine of cleaved LPXTG motif in the neighboring subunit, and the disulfide bond in the C-terminal domain (orange, orange arrowhead).
Similar articles
- The Biosynthesis and Structures of Bacterial Pili.
Lukaszczyk M, Pradhan B, Remaut H. Lukaszczyk M, et al. Subcell Biochem. 2019;92:369-413. doi: 10.1007/978-3-030-18768-2_12. Subcell Biochem. 2019. PMID: 31214993 Review. - A comprehensive guide to pilus biogenesis in Gram-negative bacteria.
Hospenthal MK, Costa TRD, Waksman G. Hospenthal MK, et al. Nat Rev Microbiol. 2017 May 12;15(6):365-379. doi: 10.1038/nrmicro.2017.40. Nat Rev Microbiol. 2017. PMID: 28496159 Review. - Assembly of complex organelles: pilus biogenesis in gram-negative bacteria as a model system.
Thanassi DG, Hultgren SJ. Thanassi DG, et al. Methods. 2000 Jan;20(1):111-26. doi: 10.1006/meth.1999.0910. Methods. 2000. PMID: 10610809 Review. - Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease.
Proft T, Baker EN. Proft T, et al. Cell Mol Life Sci. 2009 Feb;66(4):613-35. doi: 10.1007/s00018-008-8477-4. Cell Mol Life Sci. 2009. PMID: 18953686 Free PMC article. Review. - Structural biology of the chaperone-usher pathway of pilus biogenesis.
Waksman G, Hultgren SJ. Waksman G, et al. Nat Rev Microbiol. 2009 Nov;7(11):765-74. doi: 10.1038/nrmicro2220. Epub 2009 Oct 12. Nat Rev Microbiol. 2009. PMID: 19820722 Free PMC article. Review.
Cited by
- Urinary tract infections: epidemiology, mechanisms of infection and treatment options.
Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Flores-Mireles AL, et al. Nat Rev Microbiol. 2015 May;13(5):269-84. doi: 10.1038/nrmicro3432. Epub 2015 Apr 8. Nat Rev Microbiol. 2015. PMID: 25853778 Free PMC article. Review. - Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria.
Imam S, Chen Z, Roos DS, Pohlschröder M. Imam S, et al. PLoS One. 2011;6(12):e28919. doi: 10.1371/journal.pone.0028919. Epub 2011 Dec 21. PLoS One. 2011. PMID: 22216142 Free PMC article. - Genomic analysis of Indian strains of Salmonella enterica subsp. enterica serovar Typhi indicates novel genetic repertoire for pathogenicity and adaptations.
Sekhon PK, Chander AM, Mayilraj S, Rishi P. Sekhon PK, et al. Mol Biol Rep. 2019 Aug;46(4):3967-3989. doi: 10.1007/s11033-019-04843-2. Epub 2019 May 14. Mol Biol Rep. 2019. PMID: 31089918 - Structural biology of Gram-positive bacterial adhesins.
Vengadesan K, Narayana SV. Vengadesan K, et al. Protein Sci. 2011 May;20(5):759-72. doi: 10.1002/pro.613. Epub 2011 Apr 8. Protein Sci. 2011. PMID: 21404359 Free PMC article. Review. - The complete genome sequence of 'Candidatus Liberibacter solanacearum', the bacterium associated with potato zebra chip disease.
Lin H, Lou B, Glynn JM, Doddapaneni H, Civerolo EL, Chen C, Duan Y, Zhou L, Vahling CM. Lin H, et al. PLoS One. 2011 Apr 28;6(4):e19135. doi: 10.1371/journal.pone.0019135. PLoS One. 2011. PMID: 21552483 Free PMC article.
References
- Scott JR, Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. - PubMed
- Duguid JP, et al. Non-flagellar filamentous appendages (fimbriae) and haemagglutinating activity in Bacterium coli. J Pathol Bacteriol. 1955;70:335–348. - PubMed
- Yanagawa R, et al. Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Jpn J Vet Res. 1968;16:31–37. - PubMed
Publication types
MeSH terms
Grants and funding
- R37 AI048689/AI/NIAID NIH HHS/United States
- R01 AI029549/AI/NIAID NIH HHS/United States
- R01 AI048689/AI/NIAID NIH HHS/United States
- R01 AI049950/AI/NIAID NIH HHS/United States
- P50 DK064540/DK/NIDDK NIH HHS/United States
- AI49950/AI/NIAID NIH HHS/United States
- DK51406/DK/NIDDK NIH HHS/United States
- AI29549/AI/NIAID NIH HHS/United States
- DK64540/DK/NIDDK NIH HHS/United States
- AI48689/AI/NIAID NIH HHS/United States
- AI38273/AI/NIAID NIH HHS/United States
- R37 AI029549/AI/NIAID NIH HHS/United States
- R01 AI038273/AI/NIAID NIH HHS/United States
- R01 DK051406/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases