Chemical genetic inhibition of Mps1 in stable human cell lines reveals novel aspects of Mps1 function in mitosis - PubMed (original) (raw)
Chemical genetic inhibition of Mps1 in stable human cell lines reveals novel aspects of Mps1 function in mitosis
Tale Sliedrecht et al. PLoS One. 2010.
Abstract
Background: Proper execution of chromosome segregation relies on tight control of attachment of chromosomes to spindle microtubules. This is monitored by the mitotic checkpoint that allows chromosome segregation only when all chromosomes are stably attached. Proper functioning of the attachment and checkpoint processes is thus important to prevent chromosomal instability. Both processes rely on the mitotic kinase Mps1.
Principal finding: We present here two cell lines in which endogenous Mps1 has been stably replaced with a mutant kinase (Mps1-as) that is specifically inhibited by bulky PP1 analogs. Mps1 inhibition in these cell lines is highly penetrant and reversible. Timed inhibition during bipolar spindle assembly shows that Mps1 is critical for attachment error-correction and confirms its role in Aurora B regulation. We furthermore show that Mps1 has multiple controls over mitotic checkpoint activity. Mps1 inhibition precludes Mad1 localization to unattached kinetochores but also accelerates mitosis. This acceleration correlates with absence of detectable mitotic checkpoint complex after Mps1 inhibition. Finally, we show that short-term inhibition of Mps1 catalytic activity is sufficient to kill cells.
Conclusions/significance: Mps1 is involved in the regulation of multiple key processes that ensure correct chromosome segregation and is a promising target for inhibition in anti-cancer strategies. We report here two cell lines that allow specific and highly penetrant inhibition of Mps1 in a reproducible manner through the use of chemical genetics. Using these cell lines we confirm previously suggested roles for Mps1 activity in mitosis, present evidence for novel functions and examine cell viability after short and prolonged Mps1 inhibition. These cell lines present the best cellular model system to date for investigations into Mps1 biology and the effects of penetrance and duration of Mps1 inhibition on cell viability.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
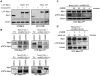
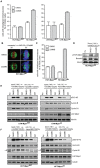
Figure 1. Stable expression of LAP-Mps1as renders selective sensitivity to 23dMB-PP1.
A: Mps1 and tubulin immunoblots of lysates of UTRM10 and HCT-TRM cells in which endogenous Mps1 is removed by doxycycline-induced expression of Mps1 shRNA and replaced with the indicated LAP-Mps1 mutants. B–D: pT676-Mps1 and Mps1 immunoblots of UTR-Mps1as (B) and HCT-Mps1as (B–D) cell lines treated with nocodazole and MG132 in combination DMSO/23dMB-PP1 for 1 hour (B), with nocodazole and MG132 for 1 hour in combination with an additional 23dMB-PP1 treatment for the indicated amounts of time (C), or with nocodazole and MG132 in combination with 23dMB-PP1 for 1 hour, followed by PBS wash after which cells were left for 15 or 30 minutes in media without 23dMB-PP1 (D).
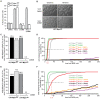
Figure 2. Mps1 activity is essential for the mitotic checkpoint.
A: Quantification of flow cytometric analysis of the fraction of mitotic cells treated with DMSO or 23dMB-PP1 alone or in combination with nocodazole for 14 hours. Graph represents average of three experiments (+/−SD). B–F: Time-lapse analysis by DIC microscopy of HCT- and UTR-derived cells treated with taxol (HCT-derived cells) or STLC (UTR-derived cells) in combination with DMSO or 23dMB-PP1. Images in B display morphology of HCT-Mps1as cells imaged at the indicated timepoints after addition of taxol in combination with DMSO or 23dMB-PP1. Graphs in C and E represent percentages of cells delayed in mitosis for 4 hours or longer. Percentages are indicated at top of each column. Line graphs in D and F show cumulative percentages of cells that display first signs of furrowing at indicated times after NEB. Dashed lines indicate half-time to furrowing of inhibited drug-treated Mps1as cells.
Figure 3. Mps1 kinase activity promotes localization of Mad1, Mad2, Bub1 and Cdc20 to unattached kinetochores.
A, B: Immunolocalization of Mad1, Mad2, Bub1, BubR1, CENP-E, Cdc20, and centromeres (ACA) in HCT-derived (A) or UTR-derived (B) cells treated with nocodazole and MG132 for 1 hour in combination with DMSO or 23dMB-PP1. Graphs represent quantifications of kinetochore signal intensities for the investigated proteins at all kinetochores in a single cell as a ratio of the ACA signal. Data are average of three experiments, 10 cells per experiment (+/− SD). Representative images are shown.
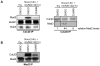
Figure 4. Mps1 inhibition reduces Mad2-Cdc20 interaction.
A, B: Immunoblots of the indicated proteins in immunoprecipitations of Cdc20 (A) or Mad2 (B). Immunoprecipitations were preformed on interphase cell lysates (thymidine, thy) or mitotic cell lysates (nocodazole, noco) of HCT-Mps1as cells treated with DMSO or 23dMB-PP1 in combination with MG132 for 1 hour. Quantification of Mad2 levels in Cdc20 immunoprecipitates, relative to interphase levels, is indicated in A, right panel.
Figure 5. Mps1 inhibition accelerates mitosis.
A–C: Time-lapse analysis by DIC microscopy of mitotic progression of HCT-derived (A, B) and UTR-derived (C) cells treated with DMSO or 23dMB-PP1. Images in A display morphology of HCT-derived cells imaged at the indicated timepoints after NEB (nuclear envelope breakdown). Graphs in B and C show cumulative percentages of cells that display first signs of furrowing at indicated times after NEB. Dashed lines indicate half-time to furrowing.
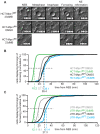
Figure 6. Mps1 activity is required for efficient correction of erroneous attachments.
A: Chromosome alignment in HCT- and UTR-Mps1WT and -Mps1as cell lines treated with MG132 for 60 minutes in combination with DMSO or 23dMB-PP1. Graphs display percentages of cells with misaligned chromosomes, averages of three experiments, at least 50 mitotic cells per experiment (+/−SD). B, C: Immunostainings of HCT-Mps1WT and HCT-Mps1as cells treated with monastrol for 30 min followed by a release from monastrol into MG132 with or without 23dMB-PP1 for 50 minutes. Cells in B were immunostained for tubulin (green) and centromeres (ACA, red). DNA (DAPI) is in blue. Graph in C indicates the percentage of mitotic cells with misaligned chromosomes 50 minutes after monastrol washout, averages of two experiments, at least 100 mitotic cells per experiment (+/−SD). D: Immunoblots of Borealin after phosphate-affinity gel electrophoresis of lysates from nocodazole-treated HCT-Mps1as cells to which DMSO or 23dMB-PP1 and MG132 was added during the final hour. E, F: Immunoblots of the indicated proteins in lysates of the UTR- (E) and HCT-Mps1 (F) variant cell lines. Cells were treated either with thymidine (thy), with taxol for 14 hours followed by the indicated inhibitors for an additional 2hrs, or with the indicated inhibitors for 1 hour followed by taxol/MG132 treatment for an additional 2 hrs ZM, ZM447439. Arrow in F points to Cyclin B. Asteriks indicates aspecific band in thymidine sample.
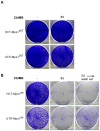
Figure 7. Specific inhibition of Mps1 enzymatic activity causes cell death.
HCT-Mps1WT and UTR-Mps1WT cells (A) and HCT-Mps1as and UTR-Mps1as cell (B) were treated with or without (-) 23dMB-PP1 for 8 days (8d), or with 23dMB-PP1 for 2 days (2d) followed by wash-out and continued growth until day 8.
Similar articles
- A Biosensor for the Mitotic Kinase MPS1 Reveals Spatiotemporal Activity Dynamics and Regulation.
Kuijt TEF, Lambers MLA, Weterings S, Ponsioen B, Bolhaqueiro ACF, Staijen DHM, Kops GJPL. Kuijt TEF, et al. Curr Biol. 2020 Oct 5;30(19):3862-3870.e6. doi: 10.1016/j.cub.2020.07.062. Epub 2020 Sep 3. Curr Biol. 2020. PMID: 32888483 - Orchestration of the spindle assembly checkpoint by CDK1-cyclin B1.
Hayward D, Alfonso-Pérez T, Gruneberg U. Hayward D, et al. FEBS Lett. 2019 Oct;593(20):2889-2907. doi: 10.1002/1873-3468.13591. Epub 2019 Sep 13. FEBS Lett. 2019. PMID: 31469407 Review. - Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine.
Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Santaguida S, et al. J Cell Biol. 2010 Jul 12;190(1):73-87. doi: 10.1083/jcb.201001036. J Cell Biol. 2010. PMID: 20624901 Free PMC article. - Mps1 regulates spindle morphology through MCRS1 to promote chromosome alignment.
Yang H, Zhang F, Huang CJ, Liao J, Han Y, Hao P, Chu Y, Lu X, Li W, Yu H, Kang J. Yang H, et al. Mol Biol Cell. 2019 Apr 15;30(9):1060-1068. doi: 10.1091/mbc.E18-09-0546. Epub 2019 Feb 20. Mol Biol Cell. 2019. PMID: 30785839 Free PMC article.
Cited by
- Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis.
Schweizer N, Ferrás C, Kern DM, Logarinho E, Cheeseman IM, Maiato H. Schweizer N, et al. J Cell Biol. 2013 Dec 23;203(6):883-93. doi: 10.1083/jcb.201309076. J Cell Biol. 2013. PMID: 24344181 Free PMC article. - Engineering and Functional Analysis of Mitotic Kinases Through Chemical Genetics.
Jones MJ, Jallepalli PV. Jones MJ, et al. Methods Mol Biol. 2016;1413:349-63. doi: 10.1007/978-1-4939-3542-0_22. Methods Mol Biol. 2016. PMID: 27193860 Free PMC article. - Kinetochore-microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint.
Etemad B, Kuijt TE, Kops GJ. Etemad B, et al. Nat Commun. 2015 Dec 1;6:8987. doi: 10.1038/ncomms9987. Nat Commun. 2015. PMID: 26621779 Free PMC article. - Characterisation of CCT271850, a selective, oral and potent MPS1 inhibitor, used to directly measure in vivo MPS1 inhibition vs therapeutic efficacy.
Faisal A, Mak GWY, Gurden MD, Xavier CPR, Anderhub SJ, Innocenti P, Westwood IM, Naud S, Hayes A, Box G, Valenti MR, De Haven Brandon AK, O'Fee L, Schmitt J, Woodward HL, Burke R, vanMontfort RLM, Blagg J, Raynaud FI, Eccles SA, Hoelder S, Linardopoulos S. Faisal A, et al. Br J Cancer. 2017 Apr 25;116(9):1166-1176. doi: 10.1038/bjc.2017.75. Epub 2017 Mar 23. Br J Cancer. 2017. PMID: 28334731 Free PMC article. - PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing.
Espert A, Uluocak P, Bastos RN, Mangat D, Graab P, Gruneberg U. Espert A, et al. J Cell Biol. 2014 Sep 29;206(7):833-42. doi: 10.1083/jcb.201406109. Epub 2014 Sep 22. J Cell Biol. 2014. PMID: 25246613 Free PMC article.
References
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8(10):798–812. - PubMed
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. - PubMed
- Kops GJPL. Dividing the goods: co-ordination of chromosome biorientation and mitotic checkpoint signalling by mitotic kinases. Biochem Soc Trans. 2009;37(Pt 5):971–975. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases