ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking - PubMed (original) (raw)
ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking
Matthew D Wilkerson et al. Bioinformatics. 2010.
Abstract
Unsupervised class discovery is a highly useful technique in cancer research, where intrinsic groups sharing biological characteristics may exist but are unknown. The consensus clustering (CC) method provides quantitative and visual stability evidence for estimating the number of unsupervised classes in a dataset. ConsensusClusterPlus implements the CC method in R and extends it with new functionality and visualizations including item tracking, item-consensus and cluster-consensus plots. These new features provide users with detailed information that enable more specific decisions in unsupervised class discovery.
Availability: ConsensusClusterPlus is open source software, written in R, under GPL-2, and available through the Bioconductor project (http://www.bioconductor.org/).
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
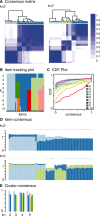
Fig. 1.
Example application of lung cancer gene expression microarrays. (A) consensus matrix, (B) item tracking plot, (C) CDF plot, (D) item-consensus plot and (E) cluster-consensus plot.
Similar articles
- DaMiRseq-an R/Bioconductor package for data mining of RNA-Seq data: normalization, feature selection and classification.
Chiesa M, Colombo GI, Piacentini L. Chiesa M, et al. Bioinformatics. 2018 Apr 15;34(8):1416-1418. doi: 10.1093/bioinformatics/btx795. Bioinformatics. 2018. PMID: 29236969 - clustComp, a bioconductor package for the comparison of clustering results.
Torrente A, Brazma A. Torrente A, et al. Bioinformatics. 2017 Dec 15;33(24):4001-4003. doi: 10.1093/bioinformatics/btx532. Bioinformatics. 2017. PMID: 28961761 Free PMC article. - mAPKL: R/ Bioconductor package for detecting gene exemplars and revealing their characteristics.
Sakellariou A, Spyrou G. Sakellariou A, et al. BMC Bioinformatics. 2015 Sep 15;16(1):291. doi: 10.1186/s12859-015-0719-5. BMC Bioinformatics. 2015. PMID: 26374744 Free PMC article. - Open source software for the analysis of microarray data.
Dudoit S, Gentleman RC, Quackenbush J. Dudoit S, et al. Biotechniques. 2003 Mar;Suppl:45-51. Biotechniques. 2003. PMID: 12664684 Review. - Big data bioinformatics.
Greene CS, Tan J, Ung M, Moore JH, Cheng C. Greene CS, et al. J Cell Physiol. 2014 Dec;229(12):1896-900. doi: 10.1002/jcp.24662. J Cell Physiol. 2014. PMID: 24799088 Free PMC article. Review.
Cited by
- Deciphering a hydrogen sulfide-related signature to supervise prognosis and therapeutic response in colon adenocarcinoma.
Chen J, Zhang Y. Chen J, et al. Medicine (Baltimore). 2024 Oct 11;103(41):e40031. doi: 10.1097/MD.0000000000040031. Medicine (Baltimore). 2024. PMID: 39465850 Free PMC article. - Expression Profile and Prognostic Significance of Pivotal Regulators for N7-Methylguanosine Methylation in Diffuse Large B-Cell Lymphoma.
Wang C, Kong R, Zhong G, Li P, Wang N, Feng G, Ding M, Zhou X. Wang C, et al. Mol Biotechnol. 2024 Oct 22. doi: 10.1007/s12033-024-01264-w. Online ahead of print. Mol Biotechnol. 2024. PMID: 39436635 - Pan-cancer characterization of metabolism-related biomarkers identifies potential therapeutic targets.
Bi G, Bian Y, Liang J, Yin J, Li R, Zhao M, Huang Y, Lu T, Zhan C, Fan H, Wang Q. Bi G, et al. J Transl Med. 2021 May 24;19(1):219. doi: 10.1186/s12967-021-02889-0. J Transl Med. 2021. PMID: 34030708 Free PMC article. - Cuproptosis-related gene index: A predictor for pancreatic cancer prognosis, immunotherapy efficacy, and chemosensitivity.
Huang X, Zhou S, Tóth J, Hajdu A. Huang X, et al. Front Immunol. 2022 Aug 25;13:978865. doi: 10.3389/fimmu.2022.978865. eCollection 2022. Front Immunol. 2022. PMID: 36090999 Free PMC article. - Characterizing m6A modification factors and their interactions in colorectal cancer: implications for tumor subtypes and clinical outcomes.
Sun W, Su Y, Zhang Z. Sun W, et al. Discov Oncol. 2024 Sep 18;15(1):457. doi: 10.1007/s12672-024-01298-1. Discov Oncol. 2024. PMID: 39292326 Free PMC article.
References
- Hayes DN, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J. Clin. Oncol. 2006;24:5079–5090. - PubMed
- Hoffmann M, et al. Optimized alignment and visualization of clustering results. In: Decker R, Lenz H.-J, editors. Advances in Data Analysis. Berlin Heidelberg: Springer; 2007. pp. 75–82.
- Monti S, et al. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 2003;52:91–118.
- Reich M, et al. GenePattern 2.0. Nat. Genet. 2006;38:500–501. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases