TumorBoost: normalization of allele-specific tumor copy numbers from a single pair of tumor-normal genotyping microarrays - PubMed (original) (raw)
TumorBoost: normalization of allele-specific tumor copy numbers from a single pair of tumor-normal genotyping microarrays
Henrik Bengtsson et al. BMC Bioinformatics. 2010.
Abstract
Background: High-throughput genotyping microarrays assess both total DNA copy number and allelic composition, which makes them a tool of choice for copy number studies in cancer, including total copy number and loss of heterozygosity (LOH) analyses. Even after state of the art preprocessing methods, allelic signal estimates from genotyping arrays still suffer from systematic effects that make them difficult to use effectively for such downstream analyses.
Results: We propose a method, TumorBoost, for normalizing allelic estimates of one tumor sample based on estimates from a single matched normal. The method applies to any paired tumor-normal estimates from any microarray-based technology, combined with any preprocessing method. We demonstrate that it increases the signal-to-noise ratio of allelic signals, making it significantly easier to detect allelic imbalances.
Conclusions: TumorBoost increases the power to detect somatic copy-number events (including copy-neutral LOH) in the tumor from allelic signals of Affymetrix or Illumina origin. We also conclude that high-precision allelic estimates can be obtained from a single pair of tumor-normal hybridizations, if TumorBoost is combined with single-array preprocessing methods such as (allele-specific) CRMA v2 for Affymetrix or BeadStudio's (proprietary) XY-normalization method for Illumina. A bounded-memory implementation is available in the open-source and cross-platform R package aroma.cn, which is part of the Aroma Project (http://www.aroma-project.org/).
Figures
Figure 1
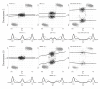
Genomic signals from genotyping microarrays in two chromosomal regions. Total (relative) copy numbers (a, e) and allele B fractions for the normal (b, f), the tumor (c, g) and the normalized tumor (d, h) for all SNPs on chromosome 2 (left) and chromosome 10 (right) in sample TCGA-23-1027. Homozygous SNPs (SNPs genotyped as AA or BB) are in gray, and heterozygous SNPs (AB) in black. Data are from the Affymetrix platform.
Figure 2
Raw paired allele B fractions. Paired observed allele B fractions, (β N, β T), of tumor TCGA-23-1027 versus its matched normal in six regions of constant PCN for the tumor. Top panels: normal (left), gained (middle), and copy-neutral LOH (right) regions from chromosome 2. Bottom panels: normal (left), deleted (middle), and copy-neutral LOH (right) regions from chromosome 10. SNPs called homozygous (AA and BB) are in gray. Linear models were robustly fitted to the heterozygous SNPs above and below the diagonal (black lines). Black discs mark the center of each cloud.
Figure 3
Paired allele B fractions after TumorBoost normalization. Paired allele B fractions, (β N,  ), and empirical densities of the raw (β T; dashed) and the normalized (
), and empirical densities of the raw (β T; dashed) and the normalized ( ; solid) allele B fractions for sample TCGA-23-1027. The same regions, SNPs and annotation as in Figure 2 are used.
; solid) allele B fractions for sample TCGA-23-1027. The same regions, SNPs and annotation as in Figure 2 are used.
Figure 4
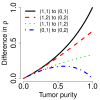
Differences in (true) decrease in heterozygosity. Differences in (true) decrease in heterozygosity (for heterozygous SNPs) between different pairs of flanking PCN regions as a function of tumor purity (κ).
Figure 5
ROC evaluation (Chr 2). (a) Left panels: The region 108.0-140.0 Mb on Chr 2 in tumor-normal sample TCGA-23-1027 has a change point at approximately 124.0 Mb, which separates a normal diploid state from a gain. 1,171 loci in each of these two states are used for the evaluation. All 79 loci in the safety region have been excluded. (b) Right panels: The region 125.0-157.0 Mb on Chr 2 in tumor-normal sample TCGA-23-1027 has a change point at approximately 141.0 Mb, which separates a normal diploid state from a gain. 986 loci in each of these two states are used for the evaluation. All 64 loci in the safety region have been excluded. The top three rows show the total CNs (C), and the raw (ρ) and normalized ( ) heterozygous DHs, respectively. A 1000 kb safety region (dashed gray frame) around the change point is excluded from the evaluation. The full resolution data points are colored black and the binned (H = 4) ones are colored blue. The three panels in the bottom row show the ROC performance of the TCNs (dotted green) and the raw (dashed black) and normalized (solid red and dot-dashed blue for naive and population-based genotypes, respectively) DHs at the full resolution (H = 1; no binning), and after binning in non-overlapping windows of size H = 2 and H = 4 SNPs, respectively.
) heterozygous DHs, respectively. A 1000 kb safety region (dashed gray frame) around the change point is excluded from the evaluation. The full resolution data points are colored black and the binned (H = 4) ones are colored blue. The three panels in the bottom row show the ROC performance of the TCNs (dotted green) and the raw (dashed black) and normalized (solid red and dot-dashed blue for naive and population-based genotypes, respectively) DHs at the full resolution (H = 1; no binning), and after binning in non-overlapping windows of size H = 2 and H = 4 SNPs, respectively.
Figure 6
ROC evaluation (Chr 10). (a) Left panels: The region 80.0-109.0 Mb on Chr 10 in tumor-normal sample TCGA-23-1027 has a change point at approximately 94.0 Mb, which separates a normal diploid state from a deletion. 1,276 loci in each of these two states are used for the evaluation. All 53 loci in the safety region have been excluded. (b) Right panels: The region 106.5-113.5 Mb on Chr 10 in tumor-normal sample TCGA-23-1027 has a change point at approximately 110.0 Mb, which separates a copy-neutral LOH region from a deletion. 254 loci in each of these two states are used for the evaluation. All 59 loci in the safety region have been excluded. The outline is the same as in Figure 5.
Figure 7
Influence of TumorBoost normalization on allele-specific copy numbers. Allele-specific CNs, (C TA, C TB), of tumor TCGA-23-1027 before (top panels) and after (bottom panels) TumorBoost normalization in a normal region (column 1), in a copy-neutral LOH region (column 2), in a gain (column 3), and in a deletion (column 4). These are some of the regions in Figure 2 using the same SNPs and annotations.
Similar articles
- CalMaTe: a method and software to improve allele-specific copy number of SNP arrays for downstream segmentation.
Ortiz-Estevez M, Aramburu A, Bengtsson H, Neuvial P, Rubio A. Ortiz-Estevez M, et al. Bioinformatics. 2012 Jul 1;28(13):1793-4. doi: 10.1093/bioinformatics/bts248. Epub 2012 May 9. Bioinformatics. 2012. PMID: 22576175 Free PMC article. - Normalization of Illumina Infinium whole-genome SNP data improves copy number estimates and allelic intensity ratios.
Staaf J, Vallon-Christersson J, Lindgren D, Juliusson G, Rosenquist R, Höglund M, Borg A, Ringnér M. Staaf J, et al. BMC Bioinformatics. 2008 Oct 2;9:409. doi: 10.1186/1471-2105-9-409. BMC Bioinformatics. 2008. PMID: 18831757 Free PMC article. - Estimation and assessment of raw copy numbers at the single locus level.
Bengtsson H, Irizarry R, Carvalho B, Speed TP. Bengtsson H, et al. Bioinformatics. 2008 Mar 15;24(6):759-67. doi: 10.1093/bioinformatics/btn016. Epub 2008 Jan 19. Bioinformatics. 2008. PMID: 18204055 - Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays.
Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, Hangaishi A, Kurokawa M, Chiba S, Gilliland DG, Koeffler HP, Ogawa S. Yamamoto G, et al. Am J Hum Genet. 2007 Jul;81(1):114-26. doi: 10.1086/518809. Epub 2007 Jun 5. Am J Hum Genet. 2007. PMID: 17564968 Free PMC article. - Single-nucleotide polymorphism array karyotyping in clinical practice: where, when, and how?
Sato-Otsubo A, Sanada M, Ogawa S. Sato-Otsubo A, et al. Semin Oncol. 2012 Feb;39(1):13-25. doi: 10.1053/j.seminoncol.2011.11.010. Semin Oncol. 2012. PMID: 22289488 Review.
Cited by
- Genetic imputation of kidney transcriptome, proteome and multi-omics illuminates new blood pressure and hypertension targets.
Xu X, Khunsriraksakul C, Eales JM, Rubin S, Scannali D, Saluja S, Talavera D, Markus H, Wang L, Drzal M, Maan A, Lay AC, Prestes PR, Regan J, Diwadkar AR, Denniff M, Rempega G, Ryszawy J, Król R, Dormer JP, Szulinska M, Walczak M, Antczak A, Matías-García PR, Waldenberger M, Woolf AS, Keavney B, Zukowska-Szczechowska E, Wystrychowski W, Zywiec J, Bogdanski P, Danser AHJ, Samani NJ, Guzik TJ, Morris AP, Liu DJ, Charchar FJ; Human Kidney Tissue Resource Study Group; Tomaszewski M. Xu X, et al. Nat Commun. 2024 Mar 19;15(1):2359. doi: 10.1038/s41467-024-46132-y. Nat Commun. 2024. PMID: 38504097 Free PMC article. - Patient-derived cells from recurrent tumors that model the evolution of _IDH_-mutant glioma.
Jones LE, Hilz S, Grimmer MR, Mazor T, Najac C, Mukherjee J, McKinney A, Chow T, Pieper RO, Ronen SM, Chang SM, Phillips JJ, Costello JF. Jones LE, et al. Neurooncol Adv. 2020 Jul 16;2(1):vdaa088. doi: 10.1093/noajnl/vdaa088. eCollection 2020 Jan-Dec. Neurooncol Adv. 2020. PMID: 32904945 Free PMC article. - A Unique Panel of Patient-Derived Cutaneous Squamous Cell Carcinoma Cell Lines Provides a Preclinical Pathway for Therapeutic Testing.
Hassan S, Purdie KJ, Wang J, Harwood CA, Proby CM, Pourreyron C, Mladkova N, Nagano A, Dhayade S, Athineos D, Caley M, Mannella V, Blyth K, Inman GJ, Leigh IM. Hassan S, et al. Int J Mol Sci. 2019 Jul 12;20(14):3428. doi: 10.3390/ijms20143428. Int J Mol Sci. 2019. PMID: 31336867 Free PMC article. - Gene Co-expression Network and Copy Number Variation Analyses Identify Transcription Factors Associated With Multiple Myeloma Progression.
Yu CY, Xiang S, Huang Z, Johnson TS, Zhan X, Han Z, Abu Zaid M, Huang K. Yu CY, et al. Front Genet. 2019 May 17;10:468. doi: 10.3389/fgene.2019.00468. eCollection 2019. Front Genet. 2019. PMID: 31156714 Free PMC article. - A comparison of per sample global scaling and per gene normalization methods for differential expression analysis of RNA-seq data.
Li X, Brock GN, Rouchka EC, Cooper NGF, Wu D, O'Toole TE, Gill RS, Eteleeb AM, O'Brien L, Rai SN. Li X, et al. PLoS One. 2017 May 1;12(5):e0176185. doi: 10.1371/journal.pone.0176185. eCollection 2017. PLoS One. 2017. PMID: 28459823 Free PMC article.
References
- Affymetrix Inc. Genome-Wide Human SNP Nsp/Sty 6.0 user guide. Affymetrix Inc; 2007. [Rev 1.]
- Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, Cheung SW, Shen RM, Barker DL, Gunderson KL. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16(9):1136–1148. doi: 10.1101/gr.5402306. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources