Sequential activation of NFAT and c-Myc transcription factors mediates the TGF-beta switch from a suppressor to a promoter of cancer cell proliferation - PubMed (original) (raw)
Sequential activation of NFAT and c-Myc transcription factors mediates the TGF-beta switch from a suppressor to a promoter of cancer cell proliferation
Garima Singh et al. J Biol Chem. 2010.
Abstract
Transforming growth factor beta (TGF-beta) has a dual role in carcinogenesis, acting as a growth inhibitor in early tumor stages and a promoter of cell proliferation in advanced diseases. Although this cellular phenomenon is well established, the underlying molecular mechanisms remain elusive. Here, we report that sequential induction of NFAT and c-Myc transcription factors is sufficient and required for the TGF-beta switch from a cell cycle inhibitor to a growth promoter pathway in cancer cells. Mechanistically, TGF-beta induces in a calcineurin-dependent manner the expression and activation of NFAT factors, which then translocate into the nucleus to promote c-Myc expression. In response to TGF-beta, activated NFAT factors bind to and displace Smad3 repressor complexes from the previously identified TGF-beta inhibitory element (TIE) to transactivate the c-Myc promoter. c-Myc in turn stimulates cell cycle progression and growth through up-regulation of D-type cyclins. Most importantly, NFAT knockdown not only prevents c-Myc activation and cell proliferation, but also partially restores TGF-beta-induced cell cycle arrest and growth suppression. Taken together, this study provides the first evidence for a Smad-independent master regulatory pathway in TGF-beta-promoted cell growth that is defined by sequential transcriptional activation of NFAT and c-Myc factors.
Figures
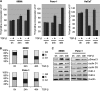
FIGURE 1.
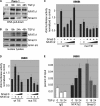
TGF-β-promoted cell cycle propagation and proliferation. A, influence of TGF-β on cell proliferation was assessed by incorporation of [3H]thymidine into Panc-1 and PaTu8988t cells after incubation in medium with 10 ng/μl TGF-β or without TGF-β for 24 and 48 h. Data are representative of triplicate experiments and are displayed as bars ± S.D. B, flow cytometry analysis was performed after propidium iodide (PI) staining in response to 10 ng/μl TGF-β treatment for 24 and 48 h. Cell cycle stages are illustrated in different colors: G2, S, and G1. Bars indicate mean values ± S.D. of three independent experiments. C, Western blot analysis to examine the effect of TGF-β on the expression of cell cycle regulatory genes in growth promoted cell lines. Panc-1 and PaTU8988t cells were left untreated or treated with TGF-β as indicated. Total cell lysates were then analyzed for expression of phosphorylated Smad3, D-type cyclins and their kinase partners (_CdK_s). Protein loading was controlled using anti-β-actin antibodies.
FIGURE 2.
Activation of TGF-β signaling induces c-Myc promoter transactivation and expression in growth promoted cells. A, TGF-β induction of c-Myc mRNA expression was analyzed by RT-PCR in Panc-1 cells. Serum-starved cells were left untreated or treated with TGF-β for 24 h before RNA extraction. mRNA expression levels were calculated relative to basal mRNA expression, which were arbitrarily set to 1 for each experiment, and expressed as fold-induction. B, induction of c-Myc was confirmed on protein levels in Panc-1 after stimulation with TGF-β for 18 and 24 h. Total cell lysates were analyzed for c-Myc protein content using an anti-c-Myc antibody. Protein loading was controlled using anti-β-actin antibodies. C, reporter gene assays illustrating the effect of TGF-β on human c-Myc promoter activity. Cells were transfected with a luciferase reporter gene construct containing the full-length wild-type c-Myc promoter sequence along with Renilla luciferase plasmids and treated with TGF-β for 18 and 24 h, respectively. Firefly luciferase reporter gene activities were measured, normalized to TK-Renilla luciferase and expressed as mean fold induction compared with untreated control that was arbitrarily set to 1. Mean values were calculated from four independent experiments and are expressed as fold induction. D, RT-PCR and Western blot analysis to demonstrate c-Myc mRNA and protein expression in HaCaT cells upon TGF-β. Serum-starved cells were left untreated or treated with TGF-β before RNA extraction or protein isolation was performed. mRNA expression levels were calculated relative to basal mRNA expression levels and expressed as fold induction. Reduction of c-Myc was confirmed on protein levels in Panc-1 after stimulation with TGF-β for 18 and 24 h. E, relevance of c-Myc induction for TGF-β induced cell proliferation was assessed by [3H]thymidine incorporation assay upon c-Myc silencing. Panc-1 cells were transfected with either control siRNA or siRNA against c-Myc. Cells were then starved and incubated in medium with or without 10 ng/μl TGF-β for 48 h. Successful c-Myc knockdown was demonstrated by immunoblotting and cell proliferation was assessed by incorporation of [3H]thymidine in control cells and in c-Myc knockdown cells as well. Bars indicate mean values ± S.D. of three independent experiments. Note that c-Myc depletion rendered cells refractory to TGF-β growth stimulation.
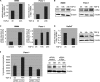
FIGURE 3.
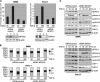
TGF-β induces NFAT factor binding to the c-Myc/TIE in cancer cells. A, cells were cotransfected with either the full-length c-Myc promoter construct (c-Myc del I, −2446 to +334) or the indicated deletions constructs (c-Myc del II-VI) or a control vector and treated with TGF-β. Firefly luciferase reporter gene activities were measured, normalized to TK-Renilla luciferase and expressed as fold induction compared with full-length c-Myc (del I). B, cells were transfected with the c-Myc/TIE reporter gene construct (−84 to −63) and TGF-β signaling was initiated through either treatment with TGF-β or co-transfection of TβR-I. Reporter gene activities were expressed as mean fold induction compared with untreated control, which was arbitrarily set to 1. Mean values were calculated from three independent experiments and are shown as mean ± S.D. C, schematic representation of the human c-Myc TIE element including previously identified binding sites for SMADs and E2F4. The GGAAA NFAT consensus sequence is also indicated. The table below displays the individual or combined mutations targeting these transcription factor binding sites as used for luciferase reporter gene assays (TIE mutants A-F). D, 8988t and Panc-1 cells were transfected with the indicated wild-type c-Myc/TIE construct (bar A) or mutant c-Myc/TIE reporter constructs (bars B–F), along with TβR-I or wild-type NFATc1/NFATc2 expression plasmids. Reporter gene activities were expressed as RLA (relative luciferase activities). Mean values were calculated from three independent experiments which were performed in triplicates. Bars indicate mean values ± S.D. E, DNA pulldown assays using double-stranded oligonucleotides of wild-type and mutant TIE with disruption of the NFAT binding site. Panc-1 cells were serum starved and then treated with TGF-β for 18, 24, or 48 h. Nuclear extracts were prepared and incubated with either wild type-TIE or TIE-NFATmut oligonucleotides. DNA-protein complexes were precipitated with streptavidin-agarose beads and NFAT binding was analyzed by Western blotting using an anti-NFATc1 antibody. F, ChIP were performed with specific NFATc1 antibodies and in vivo binding to the c-Myc promoter was determined by semi-quantitative PCR using primers specific for the c-Myc promoter region harboring the TIE element.
FIGURE 4.
NFAT expression is required for c-Myc induction by TGF-β. A, cancer cells were transfected with control siRNA or siRNA against NFAT proteins. Twenty-four hours post-transfection, cells were serum-starved and then treated with either medium alone or medium containing 10 ng/μl TGF-β. cDNA was prepared and subjected to qRT-PCR to analyze the effect of NFAT knockdown on c-Myc mRNA expression in Panc-1 (left) and PaTu8988t (right) cells. Values were calculated relative to basal mRNA expression levels in control siRNA-transfected cells, which were arbitrarily set to 1 for each experiment. Displayed are mean values from three independent experiments ± standard deviations. B, protein extracts from TGF-β-treated cancer cells transfected with either nonspecific control siRNA or NFAT-siRNA were subjected to immunoblotting with antibodies specific for NFATc1, NFATc2 or c-Myc. Protein loading was controlled using anti-β-actin antibodies.
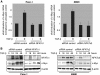
FIGURE 5.
Smad3-independent induction of NFAT proteins by TGF-β. A, Western blot analysis to show time-dependent TGF-β induction of NFAT factors in Panc-1 and PaTu8988t cells. Cells were serum-starved and then treated with TGF-β for the indicated time periods. Increased phosphorylation of Smad3 indicates successful treatment. Protein loading was controlled using anti-β-actin antibodies. B, reporter gene assays were performed in Panc-1 and PaTu8988t cells following transfection of the human NFATc1 and NFATc2 promoters and treatment with 10 ng/μl TGF-β for 18 and 24 h, respectively. Firefly luciferase reporter gene activities of the NFAT promoters were normalized to Renilla luciferase activity and expressed as RLA. Bars indicate mean values ± S.D. of three independent experiments performed in triplicates. Note that both NFAT promoters are induced by TGF-β. C, Panc-1 (left panel) and PaTu8988t (right panel) cells were transfected with siRNA against Smad3 or control siRNA, serum-starved, and treated with medium alone or medium containing TGF-β for 18, 24, or 48 h. Proteins were extracted and immunoblot analyses were performed to determine successful depletion of Smad3 and its impact on the expression levels of NFATc1, NFATc2, and c-Myc in cancer cells. Note that neither NFAT nor c-Myc expression was affected following depletion of Smad3.
FIGURE 6.
NFAT induction by TGF-β requires calcineurin phosphatase activity. A, reporter gene assays to define NFATc2 promoter regulation by Smads and CsA treatment. Cells were transfected with the human NFATc2 promoter along with either TβR-I or the Smads and incubated in the presence or absence of CsA to block endogenous calcineurin activity. Note that TGF-β induced NFATc2 promoter activation was antagonized by pharmacological inhibition of calcineurin. The NFATc2 promoter was not responsive to Smads. B, reporter gene assays to define NFATc1 promoter regulation by TGF-β and CsA. Cells were transfected with the human NFATc1 promoter and treated with either TGF-β, CsA, or a combination of both agents. Reporter gene activities were expressed as mean fold induction compared with untreated control that was arbitrarily set to 1. C, qRT-PCR to analyze the effect of CsA treatment on NFAT mRNA expression in response to TGF-β in PaTu8988t (left) and Panc-1 (right) cells. Displayed are mean values from three independent experiments ± S.D.
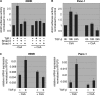
FIGURE 7.
NFAT displaces Smad3 from the c-Myc/TIE upon TGF-β. A, DNA pulldown experiment demonstrating inverse binding of NFAT factors and Smad3 on the c-Myc/TIE upon TGF-β. Nuclear extracts from Panc-1 cells were prepared and incubated with the wild-type c-Myc/TIE oligonucleotide sequence. DNA-protein complexes were precipitated with streptavidin-agarose beads, and NFAT/Smad3 binding was analyzed by Western blotting using anti-NFATc1 and anti-Smad3 antibodies, respectively. Note the inverse binding of NFAT factors and Smad3 on the c-Myc/TIE sequence upon treatment with TGF-β. B, nuclear extracts which were used for DNA pulldown experiments. C and D, reporter gene assay shows that c-Myc/TIE promoter induction depends on the integrity of the NFAT binding site and the amount of NFAT and Smad3. PaTu8988t cells were transfected with c-Myc/TIE wild type or c-Myc/TIE mutant that lacks the NFAT binding site along with increasing amounts of either NFATc2 (C) or Smad3 (D). c-Myc promoter activities were expressed as mean fold induction. Mean value were calculated from three independent experiments and are shown as mean ± S.D. E, chromatin immunoprecipitations were performed in PaTu8988 cells following TGF-β treatment over 18 and 24h using specific NFATc2 and Smad3 antibodies. In vivo binding of both transcription factors to the c-Myc promoter was determined by quantitative PCR using primers specific for the c-myc promoter region harboring the c-Myc/TIE. ChIP assay demonstrates inverse binding of Smad3 and NFAT to the c-Myc/TIE upon TGF-β.
FIGURE 8.
NFAT proteins mediate the TGF-β switch from growth suppressor to a promoter of cell proliferation. A, relevance of NFAT expression for TGF-β-induced cell proliferation was assessed by [3H]thymidine incorporation assay upon NFAT silencing. PaTu8988t and Panc-1 cells were transfected with either control siRNA or siRNA against NFAT. Cells were then starved and incubated in medium with or without 10 ng/μl TGF-β for 48 h. Bars indicate mean values ± S.D. of three independent experiments. Note that NFAT depletion rendered cells refractory to growth stimulation and partially restored TGF-β growth suppressor activities in PaTu8988T cells. Successful NFAT knockdown was demonstrated by immunoblotting using specific antibodies against NFATc1 and NFATc2 (bottom panel: control siRNA (lane 1), control siRNA + TGF-β (lane 2), siRNA NFAT (lane 3), and siRNA NFAT + TGF-β (lane 4)). B, flow cytometry analysis to study the relevance of NFAT factors in TGF-β induced cell cycle progression of cancer cells. NFAT knockdown cells were treated with TGF-β for 24 or 48 h, respectively, and analyzed by propidium iodide staining and flow cytometry. Cell cycle stages are illustrated in different colors: G2, S, and G1. Loss of NFAT expression restored cell cycle inhibition by TGF-β, as evidenced by increased cells in the G1 phase. Bars indicate mean values ± S.D. of three independent experiments. C, Western blot analysis demonstrating TGF-β regulated cell cycle genes depending on the presence or absence of NFAT expression. Cells were transfected with siRNA against NFATc2 or unspecific control siRNA, serum-starved, and subsequently treated with 10 ng/μl TGF-β for 18, 24, or 48 h. Total cell lysates were then analyzed for expression of NFAT, D-type cyclins and the partnering kinases (_CdK_s). Protein loading was controlled using anti-β-actin antibodies.
Similar articles
- NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells.
Köenig A, Linhart T, Schlengemann K, Reutlinger K, Wegele J, Adler G, Singh G, Hofmann L, Kunsch S, Büch T, Schäfer E, Gress TM, Fernandez-Zapico ME, Ellenrieder V. Köenig A, et al. Gastroenterology. 2010 Mar;138(3):1189-99.e1-2. doi: 10.1053/j.gastro.2009.10.045. Epub 2009 Nov 6. Gastroenterology. 2010. PMID: 19900447 Free PMC article. - Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway.
Buchholz M, Schatz A, Wagner M, Michl P, Linhart T, Adler G, Gress TM, Ellenrieder V. Buchholz M, et al. EMBO J. 2006 Aug 9;25(15):3714-24. doi: 10.1038/sj.emboj.7601246. Epub 2006 Jul 27. EMBO J. 2006. PMID: 16874304 Free PMC article. - Transcriptional regulation of the c-Myc promoter by NFAT1 involves negative and positive NFAT-responsive elements.
Mognol GP, de Araujo-Souza PS, Robbs BK, Teixeira LK, Viola JP. Mognol GP, et al. Cell Cycle. 2012 Mar 1;11(5):1014-28. doi: 10.4161/cc.11.5.19518. Epub 2012 Mar 1. Cell Cycle. 2012. PMID: 22333584 - TGF beta regulation of epithelial cell proliferation: role of tumor suppressor genes.
Moses HL, Pietenpol JA, Münger K, Murphy CS, Yang EY. Moses HL, et al. Princess Takamatsu Symp. 1991;22:183-95. Princess Takamatsu Symp. 1991. PMID: 1844240 Review. - The transforming growth factor beta 1/SMAD signaling pathway involved in human chronic myeloid leukemia.
Su E, Han X, Jiang G. Su E, et al. Tumori. 2010 Sep-Oct;96(5):659-66. doi: 10.1177/030089161009600503. Tumori. 2010. PMID: 21302608 Review.
Cited by
- NFAT gene family in inflammation and cancer.
Pan MG, Xiong Y, Chen F. Pan MG, et al. Curr Mol Med. 2013 May;13(4):543-54. doi: 10.2174/1566524011313040007. Curr Mol Med. 2013. PMID: 22950383 Free PMC article. Review. - NFAT3 and TGF-β/SMAD3 regulate the expression of miR-140 in osteoarthritis.
Tardif G, Pelletier JP, Fahmi H, Hum D, Zhang Y, Kapoor M, Martel-Pelletier J. Tardif G, et al. Arthritis Res Ther. 2013;15(6):R197. doi: 10.1186/ar4387. Arthritis Res Ther. 2013. PMID: 24257415 Free PMC article. - Dichotomous role of the human mitochondrial Na+/Ca2+/Li+ exchanger NCLX in colorectal cancer growth and metastasis.
Pathak T, Gueguinou M, Walter V, Delierneux C, Johnson MT, Zhang X, Xin P, Yoast RE, Emrich SM, Yochum GS, Sekler I, Koltun WA, Gill DL, Hempel N, Trebak M. Pathak T, et al. Elife. 2020 Sep 11;9:e59686. doi: 10.7554/eLife.59686. Elife. 2020. PMID: 32914752 Free PMC article. - MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies.
Hessmann E, Schneider G, Ellenrieder V, Siveke JT. Hessmann E, et al. Oncogene. 2016 Mar 31;35(13):1609-18. doi: 10.1038/onc.2015.216. Epub 2015 Jun 29. Oncogene. 2016. PMID: 26119937 Review. - NFAT transcription factors, the potion mediating "Dr. Jekill-Mr. Hyde" transformation of the TGFβ pathway in cancer cells.
Fernandez-Zapico ME, Ellenrieder V. Fernandez-Zapico ME, et al. Cell Cycle. 2010 Oct 1;9(19):3838-9. doi: 10.4161/cc.9.19.13413. Epub 2010 Oct 23. Cell Cycle. 2010. PMID: 20935481 Free PMC article. No abstract available.
References
- Chen C. R., Kang Y., Siegel P. M., Massagué J. (2002) Cell 110, 19–32 - PubMed
- Hannon G. J., Beach D. (1994) Nature 371, 257–261 - PubMed
- Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Möröy T., Bartek J., Massagué J., Hänel F., Eilers M. (2001) Nat. Cell Biol. 3, 392–399 - PubMed
- Seoane J., Pouponnot C., Staller P., Schader M., Eilers M., Massagué J. (2001) Nat. Cell Biol. 3, 400–408 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA136526/CA/NCI NIH HHS/United States
- P50CA102701/CA/NCI NIH HHS/United States
- P50 CA102701/CA/NCI NIH HHS/United States
- P30DK084567/DK/NIDDK NIH HHS/United States
- CA136526/CA/NCI NIH HHS/United States
- P30 DK084567/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous