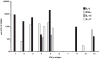Molecular determinants of T cell epitope recognition to the common Timothy grass allergen - PubMed (original) (raw)
. 2010 Jul 15;185(2):943-55.
doi: 10.4049/jimmunol.1000405. Epub 2010 Jun 16.
John Sidney, Maya F Kotturi, Ravi Kolla, Rafeul Alam, David H Broide, Stephen I Wasserman, Daniela Weiskopf, Denise M McKinney, Jo L Chung, Arnd Petersen, Howard Grey, Bjoern Peters, Alessandro Sette
Affiliations
- PMID: 20554959
- PMCID: PMC3310373
- DOI: 10.4049/jimmunol.1000405
Molecular determinants of T cell epitope recognition to the common Timothy grass allergen
Carla Oseroff et al. J Immunol. 2010.
Abstract
We investigated the molecular determinants of allergen-derived T cell epitopes in humans utilizing the Phleum pratense (Timothy grass) allergens (Phl p). PBMCs from allergic individuals were tested in ELISPOT assays with overlapping peptides spanning known Phl p allergens. A total of 43 distinct antigenic regions were recognized, illustrating the large breadth of grass-specific T cell epitopes. Th2 cytokines (as represented by IL-5) were predominant, whereas IFN-gamma, IL-10, and IL-17 were detected less frequently. Responses from specific immunotherapy treatment individuals were weaker and less consistent, yet similar in epitope specificity and cytokine pattern to allergic donors, whereas nonallergic individuals were essentially nonreactive. Despite the large breadth of recognition, nine dominant antigenic regions were defined, each recognized by multiple donors, accounting for 51% of the total response. Multiple HLA molecules and loci restricted the dominant regions, and the immunodominant epitopes could be predicted using bioinformatic algorithms specific for 23 common HLA-DR, DP, and DQ molecules. Immunodominance was also apparent at the Phl p Ag level. It was found that 52, 19, and 14% of the total response was directed to Phl p 5, 1, and 3, respectively. Interestingly, little or no correlation between Phl p-specific IgE levels and T cell responses was found. Thus, certain intrinsic features of the allergen protein might influence immunogenicity at the level of T cell reactivity. Consistent with this notion, different Phl p Ags were associated with distinct patterns of IL-5, IFN-gamma, IL-10, and IL-17 production.
Figures
FIGURE 1
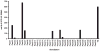
Representative pattern of CD4+ T cell cytokine secretion in response to Timothy grass-derived peptide pools. Peptides derived from Phl p 1, 2, 3, 4, 5, 6, 7, 11, 12, and 13 were synthesized as 15-mers overlapping by 10 residues and pooled in groups of ~20 peptides/pool. The distribution of CD4+ T cell secretion of IL-5 in response to these peptide pools from allergic donor U00032 is shown. Positive pools were defined as inducing a cytokine-specific response of ≥100 SFCs/106 PBMCs, p ≤ 0.05, and SI ≥ 2.0.
FIGURE 2
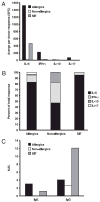
Cytokine and Ig response patterns in donor cohorts. The patterns of IL-5, IFN-γ, IL-10, and IL-17 responses in the allergic, nonallergic, and SIT donor cohorts are shown in A and B. The majority of the responses, normalized for the size of the respective cohort, was observed in allergic donors and primarily IL-5 (A). Lesser overall responses were seen in the SIT and nonallergic donor cohorts, but as with the allergics, IL-5 represented the majority of the cytokine response. B shows the percent of the total SFC response in each cohort as a function of each cytokine. Median IgE and IgG responses in the different cohorts are shown in C.
FIGURE 3
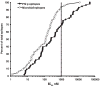
Differing distribution of HLA class II binding affinities for microbial and allergen-derived T cell epitopes. The capacity of Phl p derived epitopes to bind (IC50 nM) their respective HLA class II-restricting molecule is shown against the cumulative percent of epitopes with higher or lower affinities for their respective restricting molecule. Also shown is a similar analysis for a set of 46 HLA class II-restricted epitopes derived from various microbial sources.
FIGURE 4
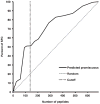
Predictions of promiscuously binding peptides cover a large fraction of the response. The capacity of each Phl p peptide to bind our panel of HLA class II molecules was predicted using a panel of algorithms as described in the text. For each peptide, the number of HLA predicted to be bound in the top 20th percentile or better was recorded. Peptides were then ranked on the basis of the number of molecules predicted to be bound. Using the number of predicted bound alleles as selection criteria, the percent of total SFCs that would have been accounted for was determined. Selecting the top fifth of peptides that bind ≥10 alleles (vertical dashed line) would have accounted for 50% of the T cell response (SFCs).
FIGURE 5
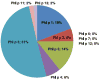
Identified T cell responses are focused on a subset of Phl p allergens. The percent fraction of total Phl p response (SFC) obtained in the cohort of allergic donors specific for each Phl p Ag is shown. The majority of the Phl p response is directed toward Phl p 5. Responses to Phl p 1 and 3 also comprise a large fraction of the overall response.
FIGURE 6
Comparison of cytokine and IgE responses in allergic donors. The relationship between total CD4+ T cell cytokine response (IL-5, IFN-γ, IL-17, and IL-10) to overlapping peptides derived from Phl p 1, 2, 3, 4, and 5 are compared with the IgE serum Ab titers against the corresponding rPhl p protein.
FIGURE 7
Cytokine response patterns vary against different Phl p allergens. The total response (SFC) specific for each cytokine and Phl p Ag was tabulated. The Phl p 1 and 11 Ags were associated with exclusive IL-5 production, whereas Phl p 4 and 5 were associated with production of all four cytokines tested. The Phl p 2, 3, and 13 Ags were associated with production of both IL-5 and IFN-γ.
Similar articles
- T-cell epitopes of Phl p 1, major pollen allergen of timothy grass (Phleum pratense): evidence for crossreacting and non-crossreacting T-cell epitopes within grass group I allergens.
Schenk S, Breiteneder H, Susani M, Najafian N, Laffer S, Duchêne M, Valenta R, Fischer G, Scheiner O, Kraft D, et al. Schenk S, et al. J Allergy Clin Immunol. 1995 Dec;96(6 Pt 1):986-96. doi: 10.1016/s0091-6749(95)70237-7. J Allergy Clin Immunol. 1995. PMID: 8543758 - Identification of isoform-specific T-cell epitopes in the major timothy grass pollen allergen, Phl p 5.
Würtzen PA, Bufe A, Wissenbach M, Madsen HO, Ipsen H, Arnved J, Van Neerven RJ. Würtzen PA, et al. Clin Exp Allergy. 1999 Dec;29(12):1614-25. doi: 10.1046/j.1365-2222.1999.00652.x. Clin Exp Allergy. 1999. PMID: 10594537 - Grass-specific CD4(+) T-cells exhibit varying degrees of cross-reactivity, implications for allergen-specific immunotherapy.
Archila LD, DeLong JH, Wambre E, James EA, Robinson DM, Kwok WW. Archila LD, et al. Clin Exp Allergy. 2014 Jul;44(7):986-98. doi: 10.1111/cea.12324. Clin Exp Allergy. 2014. PMID: 24708411 Free PMC article. - New strategies for allergen T cell epitope identification: going beyond IgE.
Schulten V, Peters B, Sette A. Schulten V, et al. Int Arch Allergy Immunol. 2014;165(2):75-82. doi: 10.1159/000368406. Epub 2014 Nov 15. Int Arch Allergy Immunol. 2014. PMID: 25402674 Free PMC article. Review. - The identification of potentially pathogenic and therapeutic epitopes from common human allergens.
Schulten V, Oseroff C, Alam R, Broide D, Vijayanand P, Peters B, Sette A. Schulten V, et al. Ann Allergy Asthma Immunol. 2013 Jan;110(1):7-10. doi: 10.1016/j.anai.2012.10.015. Epub 2012 Nov 15. Ann Allergy Asthma Immunol. 2013. PMID: 23244651 Free PMC article. Review.
Cited by
- Identification and elimination of an immunodominant T-cell epitope in recombinant immunotoxins based on Pseudomonas exotoxin A.
Mazor R, Vassall AN, Eberle JA, Beers R, Weldon JE, Venzon DJ, Tsang KY, Benhar I, Pastan I. Mazor R, et al. Proc Natl Acad Sci U S A. 2012 Dec 18;109(51):E3597-603. doi: 10.1073/pnas.1218138109. Epub 2012 Dec 3. Proc Natl Acad Sci U S A. 2012. PMID: 23213206 Free PMC article. - Ara h 1-reactive T cells in individuals with peanut allergy.
DeLong JH, Simpson KH, Wambre E, James EA, Robinson D, Kwok WW. DeLong JH, et al. J Allergy Clin Immunol. 2011 May;127(5):1211-8.e3. doi: 10.1016/j.jaci.2011.02.028. Epub 2011 Apr 2. J Allergy Clin Immunol. 2011. PMID: 21459424 Free PMC article. - Sequence conservation predicts T cell reactivity against ragweed allergens.
Pham J, Oseroff C, Hinz D, Sidney J, Paul S, Greenbaum J, Vita R, Phillips E, Mallal S, Peters B, Sette A. Pham J, et al. Clin Exp Allergy. 2016 Sep;46(9):1194-205. doi: 10.1111/cea.12772. Epub 2016 Jul 26. Clin Exp Allergy. 2016. PMID: 27359111 Free PMC article. - Peptide-induced immune regulation by a promiscuous and immunodominant CD4T-cell epitope of Timothy grass pollen: a role of Cbl-b and Itch in regulation.
Till SJ, Raynsford EJ, Reynolds CJ, Quigley KJ, Grzybowska-Kowalczyk A, Saggar LR, Goldstone A, Maillere B, Kwok WW, Altmann DM, Durham SR, Boyton RJ. Till SJ, et al. Thorax. 2014 Apr;69(4):335-45. doi: 10.1136/thoraxjnl-2013-204324. Epub 2013 Nov 20. Thorax. 2014. PMID: 24258832 Free PMC article. - Peptide binding predictions for HLA DR, DP and DQ molecules.
Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Wang P, et al. BMC Bioinformatics. 2010 Nov 22;11:568. doi: 10.1186/1471-2105-11-568. BMC Bioinformatics. 2010. PMID: 21092157 Free PMC article.
References
- Romagnani S. Regulation of the development of type 2 T-helper cells in allergy. Curr Opin Immunol. 1994;6:838–846. - PubMed
- Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. - PubMed
- Kapsenberg ML, Wierenga EA, Bos JD, Jansen HM. Functional subsets of allergen-reactive human CD4+ T cells. Immunol Today. 1991;12:392–395. - PubMed
- Romagnani S, Del Prete G, Maggi E, Parronchi P, Tiri A, Macchia D, Giudizi MG, Almerigogna F, Ricci M. Role of interleukins in induction and regulation of human IgE synthesis. Clin Immunol Immunopathol. 1989;50:S13–S23. - PubMed
- Del Prete GF, De Carli M, D’Elios MM, Maestrelli P, Ricci M, Fabbri L, Romagnani S. Allergen exposure induces the activation of allergen-specific Th2 cells in the airway mucosa of patients with allergic respiratory disorders. Eur J Immunol. 1993;23:1445–1449. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials