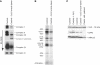Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect - PubMed (original) (raw)
Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect
Hana Antonicka et al. Am J Hum Genet. 2010.
Abstract
We investigated the genetic basis for a global and uniform decrease in mitochondrial translation in fibroblasts from patients in two unrelated pedigrees who developed Leigh syndrome, optic atrophy, and ophthalmoplegia. Analysis of the assembly of the oxidative phosphorylation complexes showed severe decreases of complexes I, IV, and V and a smaller decrease in complex III. The steady-state levels of mitochondrial mRNAs, tRNAs, and rRNAs were not reduced, nor were those of the mitochondrial translation elongation factors or the protein components of the mitochondrial ribosome. Using homozygosity mapping, we identified a 1 bp deletion in C12orf65 in one patient, and DNA sequence analysis showed a different 1 bp deletion in the second patient. Both mutations predict the same premature stop codon. C12orf65 belongs to a family of four mitochondrial class I peptide release factors, which also includes mtRF1a, mtRF1, and Ict1, all characterized by the presence of a GGQ motif at the active site. However, C12orf65 does not exhibit peptidyl-tRNA hydrolase activity in an in vitro assay with bacterial ribosomes. We suggest that it might play a role in recycling abortive peptidyl-tRNA species, released from the ribosome during the elongation phase of translation.
Copyright 2010 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
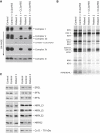
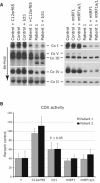
Characterization of the Molecular Defect in Patients with a Decrease in Mitochondrial Translation and Rescue of the Biochemical Phenotype by Expression of C12orf65 (A and B) Control and patient fibroblasts both alone and overexpressing C12orf65 were analyzed by BN-PAGE (A) and by pulse labeling of mitochondrial polypeptides (B). In (A), each of the five OXPHOS complexes (I–V) was visualized with a subunit-specific antibody. In (B), the seven subunits of complex I (ND), one subunit of complex III (cyt b), three subunits of complex IV (COX), and two subunits of complex V (ATP) are indicated at the left of the figure. (C) Control and patient fibroblasts were analyzed by immunoblotting with antibodies against mitochondrial translation elongation factors (EFG1, EFTu, and EFTs) and mitochondrial ribosomal proteins (MRPL13, MRPL15, MRPL32 [a kind gift of T. Langer], and MRPS2 [a kind gift of L. Spremulli, UNC Chapel Hill]). The 70 kDa subunit of complex II was used as a loading control.
Figure 2
Normal Levels of Mitochondrial mRNAs, rRNAs, and tRNAs in Patient Fibroblasts (A) RNA blot analysis was carried out with total RNA extracted from control and patient fibroblasts. Hybridization was performed with probes specific for the mitochondrial mRNAs encoding the three COX subunits, two of the complex I subunits (ND), 12S and 16S mitochondrial ribosomal RNA, and, as a loading control, a probe for beta-actin. (B) Quantification of mitochondrial mRNA and rRNA levels in fibroblasts normalized to actin levels. (C) Total RNA was extracted from control and patient cells, and 5 μg of RNA was run on a 10% polyacrylamide gel containing 7 M urea. After transfer to membrane, hybridization was performed with oligonucleotide probes complementary to the mitochondrial tRNAs for Lys, Glu, Gln, Trp, and Val, and, as a loading control, the cytosolic tRNA for Glu (cyt Glu), as indicated at the right of the figure. (D) Quantification of mitochondrial tRNA levels in patient 1 fibroblasts, normalized to levels of cytosolic tRNA for Glu.
Figure 3
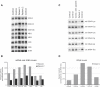
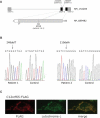
Mutational Analysis of C12orf65 and Mitochondrial Localization of the Protein (A) Schematic diagram of the C12orf65 gene and protein. The position of the mutation in the patient DNA and the position of the resulting premature stop codon are indicated. Dark boxes in the C12orf65 gene denote the coding regions; gray boxes denote 5′UTRs and 3′UTRs. The white box in C12orf65 protein indicates the predicted mitochondrial leader sequence. (B) Sequencing analysis of C12orf65 cDNA indicating the position of the homozygous 248delT and 210delA mutations in patient 1 and patient 2, respectively. (C) Control fibroblasts expressing c12orf65-FLAG were grown on coverslips and incubated with antibodies against FLAG and against cytochrome c, as indicated, followed by incubation with secondary antibodies coupled to red and green fluorescent dyes. Merging the two images demonstrates the mitochondrial localization of C12orf65.
Figure 4
Knockdown of Ict1 in Control Fibroblasts Results in a Mitochondrial Translation Defect Similar to that in Patients with C12orf65 Mutations (A and B) Control fibroblasts were transiently transfected with small interfering RNA constructs specific to Ict1 and analyzed by BN-PAGE (A) and [35S] Met/Cys-labeling (B) of mitochondrial polypeptides. (C) Control fibroblasts that were untreated, overexpressing, or depleted for Ict1 were analyzed by immunoblotting for the presence of mitochondrial ribosomal proteins (DAP3 and MRPL32). A patient with a mitochondrial ribosome assembly defect (Comb. def. patient) was used as a positive control.
Figure 5
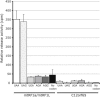
Overexpression of Ict1, but Not mtRF1 or mtRF1a/mtRF1L, Leads to a Partial Rescue of the OXPHOS Defect in C12orf65 Patients (A) BN-PAGE analysis of untreated control and patient fibroblasts and of those overexpressing C12orf65, Ict1, mtRF1, or mtRF1a/mtRF1L (mtRF1a/L). Each of the five OXPHOS complexes (I–V) was visualized with a subunit-specific antibody. (B) Quantification of COX activity (normalized to citrate synthase activity and expressed as a percentage of control) in fibroblasts from the patients alone and overexpressing C12orf65, Ict1, mtRF1, or mtRF1a/mtRF1L (3–9 replicates per sample).
Figure 6
C12orf65 Does Not Have Detectable Peptidyl-tRNA Hydrolase Activity in an In Vitro Assay with Bacterial Ribosomes E. coli ribosomes were programmed with f[3H]-Met tRNAMet at the P site and the indicated codons at the A site. Activity was measured as specific hydrolysis of f[3H]-Met from its cognate tRNAMet and expressed as cpm released. mtRF1a/mtRF1L, which recognizes the two mitochondrial termination codons UAA and UAG, was used as a positive control.
Figure 7
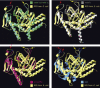
Molecular Modeling of Mitochondrial Class I Release Factor Proteins The best fit for the predicted 3D structure of C12orf65, Ict1, mtRF1, or mtRF1a/mtRF1L is the crystal structure of translation release factor RF2 from E. coli (yellow). The red arrows indicate the conserved class I release factor motif GGQ in domain III. The yellow arrows indicate the domain II loop in which the tripeptide motif (SPF in RF2) involved in stop codon recognition is positioned.
Similar articles
- A homozygous mutation of C12orf65 causes spastic paraplegia with optic atrophy and neuropathy (SPG55).
Shimazaki H, Takiyama Y, Ishiura H, Sakai C, Matsushima Y, Hatakeyama H, Honda J, Sakoe K, Naoi T, Namekawa M, Fukuda Y, Takahashi Y, Goto J, Tsuji S, Goto Y, Nakano I; Japan Spastic Paraplegia Research Consortium (JASPAC). Shimazaki H, et al. J Med Genet. 2012 Dec;49(12):777-84. doi: 10.1136/jmedgenet-2012-101212. J Med Genet. 2012. PMID: 23188110 - Structure based hypothesis of a mitochondrial ribosome rescue mechanism.
Huynen MA, Duarte I, Chrzanowska-Lightowlers ZM, Nabuurs SB. Huynen MA, et al. Biol Direct. 2012 May 8;7:14. doi: 10.1186/1745-6150-7-14. Biol Direct. 2012. PMID: 22569235 Free PMC article. - Ribosome rescue and translation termination at non-standard stop codons by ICT1 in mammalian mitochondria.
Akabane S, Ueda T, Nierhaus KH, Takeuchi N. Akabane S, et al. PLoS Genet. 2014 Sep 18;10(9):e1004616. doi: 10.1371/journal.pgen.1004616. eCollection 2014 Sep. PLoS Genet. 2014. PMID: 25233460 Free PMC article. - Optic atrophy and a Leigh-like syndrome due to mutations in the c12orf65 gene: report of a novel mutation and review of the literature.
Heidary G, Calderwood L, Cox GF, Robson CD, Teot LA, Mullon J, Anselm I. Heidary G, et al. J Neuroophthalmol. 2014 Mar;34(1):39-43. doi: 10.1097/WNO.0000000000000076. J Neuroophthalmol. 2014. PMID: 24284555 Review. - Decoding the Enigma: Translation Termination in Human Mitochondria.
Krüger A, Kovalchuk D, Shiriaev D, Rorbach J. Krüger A, et al. Hum Mol Genet. 2024 May 22;33(R1):R42-R46. doi: 10.1093/hmg/ddae032. Hum Mol Genet. 2024. PMID: 38779770 Free PMC article. Review.
Cited by
- [Clinical features and C12orf65 mutations of autosomal recessive spastic paraplegia-55: a case report].
Lin SZ, Sun XT, Ma HW. Lin SZ, et al. Zhongguo Dang Dai Er Ke Za Zhi. 2019 Nov;21(11):1094-1098. doi: 10.7499/j.issn.1008-8830.2019.11.008. Zhongguo Dang Dai Er Ke Za Zhi. 2019. PMID: 31753091 Free PMC article. Chinese. - Maternally inherited mitochondrial DNA disease in consanguineous families.
Alston CL, He L, Morris AA, Hughes I, de Goede C, Turnbull DM, McFarland R, Taylor RW. Alston CL, et al. Eur J Hum Genet. 2011 Dec;19(12):1226-9. doi: 10.1038/ejhg.2011.124. Epub 2011 Jun 29. Eur J Hum Genet. 2011. PMID: 21712854 Free PMC article. - Delineation of C12orf65-related phenotypes: a genotype-phenotype relationship.
Spiegel R, Mandel H, Saada A, Lerer I, Burger A, Shaag A, Shalev SA, Jabaly-Habib H, Goldsher D, Gomori JM, Lossos A, Elpeleg O, Meiner V. Spiegel R, et al. Eur J Hum Genet. 2014 Aug;22(8):1019-25. doi: 10.1038/ejhg.2013.284. Epub 2014 Jan 15. Eur J Hum Genet. 2014. PMID: 24424123 Free PMC article. - Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA.
Yarham JW, Lamichhane TN, Pyle A, Mattijssen S, Baruffini E, Bruni F, Donnini C, Vassilev A, He L, Blakely EL, Griffin H, Santibanez-Koref M, Bindoff LA, Ferrero I, Chinnery PF, McFarland R, Maraia RJ, Taylor RW. Yarham JW, et al. PLoS Genet. 2014 Jun 5;10(6):e1004424. doi: 10.1371/journal.pgen.1004424. eCollection 2014 Jun. PLoS Genet. 2014. PMID: 24901367 Free PMC article. - An RMND1 Mutation causes encephalopathy associated with multiple oxidative phosphorylation complex deficiencies and a mitochondrial translation defect.
Janer A, Antonicka H, Lalonde E, Nishimura T, Sasarman F, Brown GK, Brown RM, Majewski J, Shoubridge EA. Janer A, et al. Am J Hum Genet. 2012 Oct 5;91(4):737-43. doi: 10.1016/j.ajhg.2012.08.020. Epub 2012 Sep 27. Am J Hum Genet. 2012. PMID: 23022098 Free PMC article.
References
- Debray F.G., Lambert M., Mitchell G.A. Disorders of mitochondrial function. Curr. Opin. Pediatr. 2008;20:471–482. - PubMed
- Spremulli L.L., Coursey A., Navratil T., Hunter S.E. Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 2004;77:211–261. - PubMed
- Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J.E., Lochmüller H., Chevrette M., Kaufman B.A., Horvath R., Shoubridge E.A. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet. 2009;41:833–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/F011520/1/Biotechnology and Biological Sciences Research Council/United Kingdom
- Canadian Institutes of Health Research/Canada
- Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases