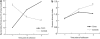Toward defining the autoimmune microbiome for type 1 diabetes - PubMed (original) (raw)
doi: 10.1038/ismej.2010.92. Epub 2010 Jul 8.
Kelsey A Gano, David B Crabb, Nabanita Mukherjee, Luis L Novelo, George Casella, Jennifer C Drew, Jorma Ilonen, Mikael Knip, Heikki Hyöty, Riitta Veijola, Tuula Simell, Olli Simell, Josef Neu, Clive H Wasserfall, Desmond Schatz, Mark A Atkinson, Eric W Triplett
Affiliations
- PMID: 20613793
- PMCID: PMC3105672
- DOI: 10.1038/ismej.2010.92
Toward defining the autoimmune microbiome for type 1 diabetes
Adriana Giongo et al. ISME J. 2011 Jan.
Abstract
Several studies have shown that gut bacteria have a role in diabetes in murine models. Specific bacteria have been correlated with the onset of diabetes in a rat model. However, it is unknown whether human intestinal microbes have a role in the development of autoimmunity that often leads to type 1 diabetes (T1D), an autoimmune disorder in which insulin-secreting pancreatic islet cells are destroyed. High-throughput, culture-independent approaches identified bacteria that correlate with the development of T1D-associated autoimmunity in young children who are at high genetic risk for this disorder. The level of bacterial diversity diminishes overtime in these autoimmune subjects relative to that of age-matched, genotype-matched, nonautoimmune individuals. A single species, Bacteroides ovatus, comprised nearly 24% of the total increase in the phylum Bacteroidetes in cases compared with controls. Conversely, another species in controls, represented by the human firmicute strain CO19, represented nearly 20% of the increase in Firmicutes compared with cases overtime. Three lines of evidence are presented that support the notion that, as healthy infants approach the toddler stage, their microbiomes become healthier and more stable, whereas, children who are destined for autoimmunity develop a microbiome that is less diverse and stable. Hence, the autoimmune microbiome for T1D may be distinctly different from that found in healthy children. These data also suggest bacterial markers for the early diagnosis of T1D. In addition, bacteria that negatively correlated with the autoimmune state may prove to be useful in the prevention of autoimmunity development in high-risk children.
Figures
Figure 1
Significant differences in taxa between cases (autoimmune) and controls (healthy). Samples were collected approximately 4 months, 1 year and 2 years after birth, represented, respectively, as time points 1, 2 and 3: (a) increasing numbers of Bacteroidetes in cases overtime compared with controls; (b) increasing numbers of Firmicutes in controls overtime compared with cases; and (c) higher proportion of unclassified sequences in controls compared with cases. Significant differences between cases and controls are designated by a star (_P_⩽0.002).
Figure 2
Bacterial community differences between cases and controls during autoimmunity development in cases; (a) significant increase in Bacteroidetes with concomitant decrease in Firmicutes in cases compared with controls (_P_-value ⩽0.01 at all time points; (b) significantly higher (P<0.05) Shannon diversity index in controls compared with cases in time point 3. Significant differences between cases and controls are designated by a star. The _P_-values for time points 1, 2 and 3 are (a) 0.0000, 0.0000 and 0.0000, and (b) 0.80, 0.33 and 0.03, respectively.
Figure 3
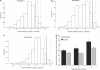
Histograms showing the permutation test based on the UniFrac significance obtained from the three time points (a, b and c). Dashed blue lines represent the 0.10, 0.05 and 0.01 quantiles, and the red line indicates the value of the observed difference. No differences in community diversity were observed at time point 1 at the 10% confidence interval. However, the average distance between any pair of cases was significantly higher than that between any pair of controls at the second and third collection points at the 5% and 10% level of confidence, respectively. A summary of data over all time points is shown in (d). (The color version of this figure is available in online version only).
Figure 4
Principal coordinate analysis for the case and control communities at time points 1 (a), 2 (b) and 3 (c).
Similar articles
- Alterations in Intestinal Microbiota Correlate With Susceptibility to Type 1 Diabetes.
Alkanani AK, Hara N, Gottlieb PA, Ir D, Robertson CE, Wagner BD, Frank DN, Zipris D. Alkanani AK, et al. Diabetes. 2015 Oct;64(10):3510-20. doi: 10.2337/db14-1847. Epub 2015 Jun 11. Diabetes. 2015. PMID: 26068542 Free PMC article. - The human gut microbiome in early-onset type 1 diabetes from the TEDDY study.
Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, Lernmark Å, Hagopian WA, Rewers MJ, She JX, Toppari J, Ziegler AG, Akolkar B, Krischer JP, Stewart CJ, Ajami NJ, Petrosino JF, Gevers D, Lähdesmäki H, Vlamakis H, Huttenhower C, Xavier RJ. Vatanen T, et al. Nature. 2018 Oct;562(7728):589-594. doi: 10.1038/s41586-018-0620-2. Epub 2018 Oct 24. Nature. 2018. PMID: 30356183 Free PMC article. - Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes.
Maffeis C, Martina A, Corradi M, Quarella S, Nori N, Torriani S, Plebani M, Contreas G, Felis GE. Maffeis C, et al. Diabetes Metab Res Rev. 2016 Oct;32(7):700-709. doi: 10.1002/dmrr.2790. Epub 2016 Mar 30. Diabetes Metab Res Rev. 2016. PMID: 26891226 - Impact of type 1 diabetes on the composition and functional potential of gut microbiome in children and adolescents: possible mechanisms, current knowledge, and challenges.
Mokhtari P, Metos J, Anandh Babu PV. Mokhtari P, et al. Gut Microbes. 2021 Jan-Dec;13(1):1-18. doi: 10.1080/19490976.2021.1926841. Gut Microbes. 2021. PMID: 34101547 Free PMC article. Review. - Type 1 Diabetes Mellitus and Celiac Disease: Distinct Autoimmune Disorders That Share Common Pathogenic Mechanisms.
Goodwin G. Goodwin G. Horm Res Paediatr. 2019;92(5):285-292. doi: 10.1159/000503142. Epub 2019 Oct 8. Horm Res Paediatr. 2019. PMID: 31593953 Review.
Cited by
- Regulatory properties of the intestinal microbiome effecting the development and treatment of diabetes.
Romano-Keeler J, Weitkamp JH, Moore DJ. Romano-Keeler J, et al. Curr Opin Endocrinol Diabetes Obes. 2012 Apr;19(2):73-80. doi: 10.1097/MED.0b013e3283514d43. Curr Opin Endocrinol Diabetes Obes. 2012. PMID: 22357099 Free PMC article. Review. - Risk factors for type 1 diabetes, including environmental, behavioural and gut microbial factors: a case-control study.
Traversi D, Rabbone I, Scaioli G, Vallini C, Carletto G, Racca I, Ala U, Durazzo M, Collo A, Ferro A, Carrera D, Savastio S, Cadario F, Siliquini R, Cerutti F. Traversi D, et al. Sci Rep. 2020 Oct 16;10(1):17566. doi: 10.1038/s41598-020-74678-6. Sci Rep. 2020. PMID: 33067559 Free PMC article. - The microbiology of human hygiene and its impact on type 1 diabetes.
Chapman NM, Coppieters K, von Herrath M, Tracy S. Chapman NM, et al. Islets. 2012 Jul-Aug;4(4):253-61. doi: 10.4161/isl.21570. Epub 2012 Jul 1. Islets. 2012. PMID: 22996796 Free PMC article. Review. - A model for the role of gut bacteria in the development of autoimmunity for type 1 diabetes.
Davis-Richardson AG, Triplett EW. Davis-Richardson AG, et al. Diabetologia. 2015 Jul;58(7):1386-93. doi: 10.1007/s00125-015-3614-8. Epub 2015 May 10. Diabetologia. 2015. PMID: 25957231 Free PMC article. Review. - Nutri(meta)genetics and cardiovascular disease: novel concepts in the interaction of diet and genomic variation.
Joseph J, Loscalzo J. Joseph J, et al. Curr Atheroscler Rep. 2015 May;17(5):505. doi: 10.1007/s11883-015-0505-x. Curr Atheroscler Rep. 2015. PMID: 25782777 Free PMC article. Review.
References
- Brugman S, Klatter FA, Visser JTJ, Wildeboer-Veloo ACM, Harmsen HJM, Rozing J, et al. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes. Diabetologia. 2006;49:2105–2108. - PubMed
- Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S, et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia. 2005;48:1565–1575. - PubMed
- de Kruif P. Microbe Hunters. Harcourt, Brace and Co.: New York; 1926.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical