The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data - PubMed (original) (raw)
The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data
Aaron McKenna et al. Genome Res. 2010 Sep.
Abstract
Next-generation DNA sequencing (NGS) projects, such as the 1000 Genomes Project, are already revolutionizing our understanding of genetic variation among individuals. However, the massive data sets generated by NGS--the 1000 Genome pilot alone includes nearly five terabases--make writing feature-rich, efficient, and robust analysis tools difficult for even computationally sophisticated individuals. Indeed, many professionals are limited in the scope and the ease with which they can answer scientific questions by the complexity of accessing and manipulating the data produced by these machines. Here, we discuss our Genome Analysis Toolkit (GATK), a structured programming framework designed to ease the development of efficient and robust analysis tools for next-generation DNA sequencers using the functional programming philosophy of MapReduce. The GATK provides a small but rich set of data access patterns that encompass the majority of analysis tool needs. Separating specific analysis calculations from common data management infrastructure enables us to optimize the GATK framework for correctness, stability, and CPU and memory efficiency and to enable distributed and shared memory parallelization. We highlight the capabilities of the GATK by describing the implementation and application of robust, scale-tolerant tools like coverage calculators and single nucleotide polymorphism (SNP) calling. We conclude that the GATK programming framework enables developers and analysts to quickly and easily write efficient and robust NGS tools, many of which have already been incorporated into large-scale sequencing projects like the 1000 Genomes Project and The Cancer Genome Atlas.
Figures
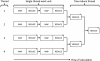
Figure 1.
Read-based and locus-based traversals. Read-based traversals provide a sequencer read and its associated reference data during each iteration of the traversal. Locus-based traversals are provided with the reference base, associated reference ordered data, and the pileup of read bases at the given locus. These iterations are repeated respectively for each read or each reference base in the input BAM file.
Figure 2.
Shared memory parallel tree-reduction in the GATK. Each thread executes independent MapReduce calls on a single instance of the analysis walker, and the GATK uses the user specified tree-reduce function to merge together the reduce results of each thread in sequential order. The final in-order reduce result is returned.
Figure 3.
MHC depth of coverage in JPT samples of the 1000 Genomes Project pilot 2, calculated using the GATK depth of coverage tool. Coverage is averaged over 2.5-kb regions, where lines represent a local polynomial regression of coverage. The track containing all known annotated genes from the UCSC Genome Browser is shown in gray, with HLA genes highlighted in red. Coverage drops near 32.1 M and 32.7 M correspond with increasing density of HLA genes.
Figure 4.
Code sample for the simple genotyper walker. The map function uses a naïve Bayesian method to generate genotypes, given the pileup of reference bases at the current locus, and emits a call containing the likelihoods for each of the 10 possible genotypes (assuming a diploid organism). This is then output to disk. The implementation of the tree-reduce function provides directions to the GATK engine for reducing two in-order parallel reduce results, allowing parallelization of the genotyper.
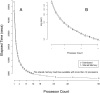
Figure 5.
Parallelization of genotyping in the GATK. (A) 1000 Genomes Project sample NA12878s chromosome 1 was genotyped using both shared memory parallelization and distributed parallelization methods. Both methods follow a near exponential curve (B) as the processor count was increased, and using the distributed methodology it was possible to see elapsed time gains out to 50 processors.
Similar articles
- Optimizing performance of GATK workflows using Apache Arrow In-Memory data framework.
Ahmad T, Ahmed N, Al-Ars Z, Hofstee HP. Ahmad T, et al. BMC Genomics. 2020 Nov 18;21(Suppl 10):683. doi: 10.1186/s12864-020-07013-y. BMC Genomics. 2020. PMID: 33208101 Free PMC article. - PhredEM: a phred-score-informed genotype-calling approach for next-generation sequencing studies.
Liao P, Satten GA, Hu YJ. Liao P, et al. Genet Epidemiol. 2017 Jul;41(5):375-387. doi: 10.1002/gepi.22048. Epub 2017 May 31. Genet Epidemiol. 2017. PMID: 28560825 Free PMC article. - OVarFlow: a resource optimized GATK 4 based Open source Variant calling workFlow.
Bathke J, Lühken G. Bathke J, et al. BMC Bioinformatics. 2021 Aug 13;22(1):402. doi: 10.1186/s12859-021-04317-y. BMC Bioinformatics. 2021. PMID: 34388963 Free PMC article. - Structural variation discovery in the cancer genome using next generation sequencing: computational solutions and perspectives.
Liu B, Conroy JM, Morrison CD, Odunsi AO, Qin M, Wei L, Trump DL, Johnson CS, Liu S, Wang J. Liu B, et al. Oncotarget. 2015 Mar 20;6(8):5477-89. doi: 10.18632/oncotarget.3491. Oncotarget. 2015. PMID: 25849937 Free PMC article. Review. - Current state-of-art of sequencing technologies for plant genomics research.
Thudi M, Li Y, Jackson SA, May GD, Varshney RK. Thudi M, et al. Brief Funct Genomics. 2012 Jan;11(1):3-11. doi: 10.1093/bfgp/elr045. Brief Funct Genomics. 2012. PMID: 22345601 Review.
Cited by
- Whole-exome sequencing reveals genomic landscape of intrahepatic cholangiocarcinoma and identifies SAV1 as a potential driver.
Zhou ZJ, Ye YH, Hu ZQ, Hou YR, Liu KX, Sun RQ, Wang PC, Luo CB, Li J, Zou JX, Zhou J, Fan J, Song CL, Zhou SL. Zhou ZJ, et al. Nat Commun. 2024 Nov 17;15(1):9960. doi: 10.1038/s41467-024-54387-8. Nat Commun. 2024. PMID: 39551842 - A Lassa virus live attenuated vaccine candidate that is safe and efficacious in guinea pigs.
Carey BD, Yu S, Geiger J, Ye C, Huzella LM, Reeder RJ, Mehta M, Hirsch S, Bernbaum R, Cubitt B, Pahar B, Anthony SM, Marketon A, Bernbaum JG, Tran JP, Crozier I, Martínez-Sobrido L, Worwa G, de la Torre JC, Kuhn JH. Carey BD, et al. NPJ Vaccines. 2024 Nov 17;9(1):220. doi: 10.1038/s41541-024-01012-w. NPJ Vaccines. 2024. PMID: 39551823 - MGA loss-of-function variants cause premature ovarian insufficiency.
Tang S, Guo T, Song C, Wang L, Zhang J, Rajkovic A, Lin X, Chen S, Liu Y, Tian W, Wu B, Wang S, Wang W, Lai Y, Wang A, Xu S, Jin L, Ke H, Zhao S, Li Y, Qin Y, Zhang F, Chen ZJ. Tang S, et al. J Clin Invest. 2024 Nov 15;134(22):e183758. doi: 10.1172/JCI183758. J Clin Invest. 2024. PMID: 39545409 Free PMC article. - Whole-genome sequencing reveals genetic structure and adaptive genes in Nepalese buffalo breeds.
Dhakal A, Si J, Sapkota S, Pauciullo A, Han J, Gorkhali NA, Zhao X, Zhang Y. Dhakal A, et al. BMC Genomics. 2024 Nov 14;25(1):1082. doi: 10.1186/s12864-024-10993-w. BMC Genomics. 2024. PMID: 39543523 Free PMC article. - Identification and validation of two quantitative trait loci showing pleiotropic effect on multiple grain-related traits in bread wheat (Triticum aestivum L.).
Hu W, You J, Yong R, Zhao D, Li D, Wang Z, Jia J. Hu W, et al. Theor Appl Genet. 2024 Nov 14;137(12):268. doi: 10.1007/s00122-024-04778-8. Theor Appl Genet. 2024. PMID: 39540955
References
- Bhandarkar M 2009. Practical problem solving with hadoop and pig. In USENIX. The USENIX Association, San Diego, CA
- Dean J, Ghemawat S 2008. MapReduce: Simplified data processing on large clusters. Commun ACM 51: 107–113
Publication types
MeSH terms
Grants and funding
- P30 DK043351/DK/NIDDK NIH HHS/United States
- U01 HG005208/HG/NHGRI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- 54 HG003067/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources