Lung cancer cell lines as tools for biomedical discovery and research - PubMed (original) (raw)
Review
. 2010 Sep 8;102(17):1310-21.
doi: 10.1093/jnci/djq279. Epub 2010 Aug 2.
Affiliations
- PMID: 20679594
- PMCID: PMC2935474
- DOI: 10.1093/jnci/djq279
Review
Lung cancer cell lines as tools for biomedical discovery and research
Adi F Gazdar et al. J Natl Cancer Inst. 2010.
Abstract
Lung cancer cell lines have made a substantial contribution to lung cancer translational research and biomedical discovery. A systematic approach to initiating and characterizing cell lines from small cell and non-small cell lung carcinomas has led to the current collection of more than 200 lung cancer cell lines, a number that exceeds those for other common epithelial cancers combined. The ready availability and widespread dissemination of the lines to investigators worldwide have resulted in more than 9000 citations, including multiple examples of important biomedical discoveries. The high (but not perfect) genomic similarities between lung cancer cell lines and the lung tumor type from which they were derived provide evidence of the relevance of their use. However, major problems including misidentification or cell line contamination remain. Ongoing studies and new approaches are expected to reveal the full potential of the lung cancer cell line panel.
Figures
Figure 1
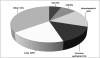
Distribution of human tumor cell lines in the Sanger database (
). Of the 784 cell lines for which data were available, 154 were of lung origin. The common epithelial category group includes breast, colorectal, prostate, gastric, and hepatic cancers. The “other” group includes all other types of human cancers listed in the database. CNS = central nervous system.
Figure 2
Regions of frequent amplification and deletion in lung adenocarcinoma cell lines and tumors. A) Alteration frequency. Thirty-three lung adenocarcinoma cell lines and 169 tumor samples were analyzed by use of a whole-genome tiling path array for comparative genomic hybridization analysis (33). Genomic imbalances were identified by use of aCGH-Smooth (34), and the frequency of alteration across all samples in each group was determined as described previously (35). The resulting alteration frequencies are displayed across the entire human genome with amplifications on the left and deletions on the right. For each chromosome, the shaded portion represents the p arm. Chromosome location is depicted on the left of the histograms with the p and q arms separated by differential shading (gray and white, respectively). The genomic similarity between the cell lines and tumors is evident by the close association of the plots. B) Commonly reported sites of amplification and deletion that are frequently altered (>15% of samples) in both cell line and tumor samples. Frequency values for chromosome bands were generated as described above. Known oncogenes and tumor suppressor genes that are located in each region are shown.
Figure 3
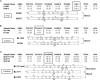
Verification of the identity of lung cancer cell lines. Representative electropherograms from PowerPlex fingerprint analyses are shown that verify the identity of cell lines from different sources. For sample analysis, the allele sizes of the nine polymorphic markers analyzed are displayed below the electropherograms. The electropherogram results of one marker (boxed and also identified to the left) are displayed. When at least seven of the nine markers matched, the cell lines are assumed to be identical. A) Match of cell lines H1993 and H2073. Both cell lines were isolated from the same patient and, thus, this represents a match. B) Match of B lymphoblastoid cell line BL1184 and non–small cell lung cancer cell line H1184. Both cell lines were isolated from the same patient. C) Incorrect attribution. BL1171 cells (middle) match BL2195 cells as a result of cell line cross-contamination or mislabeling. Note that full designations of the cell lines include the prefix NCI (eg, NCI-BL1171).
Figure 4
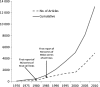
Number of citations for human lung cancer cell lines. Data were obtained from a search of the PubMed database from within the EndNote program by use of the medical search heading terms “lung neoplasms”; “cell line, tumor”; and “humans.” The cumulative number of citations and the number of citations per 5 years are shown. The results are from a search performed in June 2010. NCI = National Cancer Institute; NSCLC = non–small cell lung cancer; SCLC = small cell lung cancer.
Similar articles
- [Anti-angiogenic factors in thoracic oncology: successes, failures and prospects].
Marco S, Tomasini P, Greillier L, Barlesi F. Marco S, et al. Rev Mal Respir. 2011 Dec;28(10):1216-29. doi: 10.1016/j.rmr.2011.04.014. Epub 2011 Nov 4. Rev Mal Respir. 2011. PMID: 22152931 Review. French. - Comprehensive Pharmacogenomic Profiling of Malignant Pleural Mesothelioma Identifies a Subgroup Sensitive to FGFR Inhibition.
Quispel-Janssen JM, Badhai J, Schunselaar L, Price S, Brammeld J, Iorio F, Kolluri K, Garnett M, Berns A, Baas P, McDermott U, Neefjes J, Alifrangis C. Quispel-Janssen JM, et al. Clin Cancer Res. 2018 Jan 1;24(1):84-94. doi: 10.1158/1078-0432.CCR-17-1172. Epub 2017 Oct 23. Clin Cancer Res. 2018. PMID: 29061644 - Human lung-derived mesenchymal stem cell-conditioned medium exerts in vitro antitumor effects in malignant pleural mesothelioma cell lines.
Cortes-Dericks L, Froment L, Kocher G, Schmid RA. Cortes-Dericks L, et al. Stem Cell Res Ther. 2016 Feb 9;7:25. doi: 10.1186/s13287-016-0282-7. Stem Cell Res Ther. 2016. PMID: 26861734 Free PMC article. - Eve of the third millennium, providing an update on the management of non-small cell lung cancer (NSCLC).
Le Chevalier T. Le Chevalier T. Anticancer Drugs. 2001 Jul;12 Suppl 3:S1. Anticancer Drugs. 2001. PMID: 11556248 English, French. No abstract available. - Antibody drug conjugates in thoracic malignancies.
Pacheco JM, Camidge DR. Pacheco JM, et al. Lung Cancer. 2018 Oct;124:260-269. doi: 10.1016/j.lungcan.2018.07.001. Epub 2018 Jul 19. Lung Cancer. 2018. PMID: 30268471 Review.
Cited by
- Mode of action and pharmacogenomic biomarkers for exceptional responders to didemnin B.
Potts MB, McMillan EA, Rosales TI, Kim HS, Ou YH, Toombs JE, Brekken RA, Minden MD, MacMillan JB, White MA. Potts MB, et al. Nat Chem Biol. 2015 Jun;11(6):401-8. doi: 10.1038/nchembio.1797. Epub 2015 Apr 13. Nat Chem Biol. 2015. PMID: 25867045 Free PMC article. - Modulation of the cancer cell transcriptome by culture media formulations and cell density.
Kim SW, Kim SJ, Langley RR, Fidler IJ. Kim SW, et al. Int J Oncol. 2015 May;46(5):2067-75. doi: 10.3892/ijo.2015.2930. Epub 2015 Mar 17. Int J Oncol. 2015. PMID: 25776572 Free PMC article. - PLCγ1 suppression promotes the adaptation of KRAS-mutant lung adenocarcinomas to hypoxia.
Saliakoura M, Rossi Sebastiano M, Pozzato C, Heidel FH, Schnöder TM, Savic Prince S, Bubendorf L, Pinton P, A Schmid R, Baumgartner J, Freigang S, Berezowska SA, Rimessi A, Konstantinidou G. Saliakoura M, et al. Nat Cell Biol. 2020 Nov;22(11):1382-1395. doi: 10.1038/s41556-020-00592-8. Epub 2020 Oct 19. Nat Cell Biol. 2020. PMID: 33077911 Free PMC article. - Drug sensitivity in cancer cell lines is not tissue-specific.
Jaeger S, Duran-Frigola M, Aloy P. Jaeger S, et al. Mol Cancer. 2015 Feb 15;14:40. doi: 10.1186/s12943-015-0312-6. Mol Cancer. 2015. PMID: 25881072 Free PMC article. - Stromal platelet-derived growth factor receptor α (PDGFRα) provides a therapeutic target independent of tumor cell PDGFRα expression in lung cancer xenografts.
Gerber DE, Gupta P, Dellinger MT, Toombs JE, Peyton M, Duignan I, Malaby J, Bailey T, Burns C, Brekken RA, Loizos N. Gerber DE, et al. Mol Cancer Ther. 2012 Nov;11(11):2473-82. doi: 10.1158/1535-7163.MCT-12-0431. Epub 2012 Aug 28. Mol Cancer Ther. 2012. PMID: 22933705 Free PMC article.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. - PubMed
- Weed D. Methodologic guidelines for review papers. J Natl Cancer Inst. 1997;89(1):6–7.
- Lucey BP, Nelson-Rees WA, Hutchins GM. Henrietta Lacks, HeLa cells, and cell culture contamination. Arch Pathol Lab Med. 2009;133(9):1463–1467. - PubMed
- Nelson-Rees WA, Zhdanov VM, Hawthorne PK, Flandermeyer RR. HeLa-like marker chromosomes and type-A variant glucose-6-phosphate dehydrogenase isoenzyme in human cell cultures producing Mason-Pfizer monkey virus-like particles. J Natl Cancer Inst. 1974;53(3):751–757. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials