Musite, a tool for global prediction of general and kinase-specific phosphorylation sites - PubMed (original) (raw)
Musite, a tool for global prediction of general and kinase-specific phosphorylation sites
Jianjiong Gao et al. Mol Cell Proteomics. 2010 Dec.
Abstract
Reversible protein phosphorylation is one of the most pervasive post-translational modifications, regulating diverse cellular processes in various organisms. High throughput experimental studies using mass spectrometry have identified many phosphorylation sites, primarily from eukaryotes. However, the vast majority of phosphorylation sites remain undiscovered, even in well studied systems. Because mass spectrometry-based experimental approaches for identifying phosphorylation events are costly, time-consuming, and biased toward abundant proteins and proteotypic peptides, in silico prediction of phosphorylation sites is potentially a useful alternative strategy for whole proteome annotation. Because of various limitations, current phosphorylation site prediction tools were not well designed for comprehensive assessment of proteomes. Here, we present a novel software tool, Musite, specifically designed for large scale predictions of both general and kinase-specific phosphorylation sites. We collected phosphoproteomics data in multiple organisms from several reliable sources and used them to train prediction models by a comprehensive machine-learning approach that integrates local sequence similarities to known phosphorylation sites, protein disorder scores, and amino acid frequencies. Application of Musite on several proteomes yielded tens of thousands of phosphorylation site predictions at a high stringency level. Cross-validation tests show that Musite achieves some improvement over existing tools in predicting general phosphorylation sites, and it is at least comparable with those for predicting kinase-specific phosphorylation sites. In Musite V1.0, we have trained general prediction models for six organisms and kinase-specific prediction models for 13 kinases or kinase families. Although the current pretrained models were not correlated with any particular cellular conditions, Musite provides a unique functionality for training customized prediction models (including condition-specific models) from users' own data. In addition, with its easily extensible open source application programming interface, Musite is aimed at being an open platform for community-based development of machine learning-based phosphorylation site prediction applications. Musite is available at http://musite.sourceforge.net/.
Figures
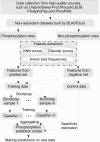
Fig. 1.
Overall work flow of Musite.
Fig. 2.
Comparison of KNN scores between phosphorylation sites and non-phosphorylation sites. KNN scores of 1,000 phosphorylation sites and 1,000 non-phosphorylation sites randomly selected from each non-redundant data sets for six organisms were plotted. A, box plots of KNN scores (H. sapiens serine/threonine data only) for phosphorylation sites (red) and non-phosphorylation sites (blue). The horizontal axis represents the size of nearest neighbors (in percentage of the bootstrapped data set size). The vertical axis represents the KNN score. The bottom and top of the box are the 25th and 75th percentiles, respectively; the central band is the median; the whiskers extend to the most extreme data points that are not considered outliers; and the outliers are plotted individually as plus marks (+). B, comparison of mean KNN scores between phosphorylation sites (pentagrams) and non-phosphorylation sites (circles) in six organisms.
Fig. 3.
Preference of phosphorylation sites in disordered regions. Disorder scores for the H. sapiens NR data set and the A. thaliana NR data set are shown as examples. All phosphorylation sites and non-phosphorylation sites that have 6 or more residues at both sides were used. A, histogram of disorder scores of residues around phosphoserines/threonines (23,907 in total) in the H. sapiens NR data set. The horizontal axis represents the disorder score predicted by VSL2B, divided evenly into 10 subranges from 0 to 1; the vertical axis represents the occurrence (the number of sites) in the corresponding disorder subrange. Different colors from blue to red in each bar stand for 13 different residue positions in the window from the upstream −6 to downstream +6 residues as indicated in the color bar on the right. B, histogram of disorder scores of residues around non-phosphoserines/threonines (1,171,139 in total) in the H. sapiens NR data set. C, histogram of disorder scores of residues around phosphoserine/threonine sites (3,512 in total) in the A. thaliana NR data set. D, histogram of disorder scores of residues around non-phosphoserine/threonine sites (986,481 in total) in the A. thaliana NR data set. E, histogram of disorder scores of residues around phosphotyrosine sites (2,504 in total) in the H. sapiens NR data set. F, histogram of disorder scores of residues around non-phosphotyrosine sites (221,322 in total) in the H. sapiens NR data set.
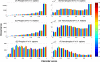
Fig. 4.
Comparisons of amino acid compositions in positive and negative data sets. A, comparisons between phosphoserines/threonines and non-phosphoserines/threonines in six organisms. The vertical axis represents the log2 ratio between amino acid frequencies surrounding phosphoserines/threonines and those surrounding non-phosphoserines/threonines. A value larger than 0 means the corresponding amino acid is enriched surrounding phosphoserines/threonines. The horizontal axis represents the 20 amino acids sorted in descending order by the mean log2 ratio. B, similarly, comparisons between phosphotyrosines and non-phosphotyrosines in H. sapiens and M. musculus (phosphotyrosine data in the other four organisms are too sparse to derive meaningful statistics).
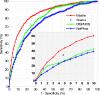
Fig. 5.
ROC curves of Musite predictions on NR data sets of H. sapiens, M. musculus, D. melanogaster, C. elegans, S. cerevisiae, and A. thaliana. Each curve represents the average sensitivities and specificities for difference thresholds over 10 cross-validation runs. The bottom right figure is the zoomed-in region with high prediction specificities (0.9–1).
Fig. 6.
Comparison of phosphoserine/threonine prediction performances of NetPhos, DISPHOS, scan-x, and Musite. For NetPhos, DISPHOS, and Musite, the phosphoserine/threonine prediction scores were extracted, and the corresponding ROC curves were calculated and plotted. For scan-x, only specificities/sensitivities at the two supported stringency levels were plotted. The bottom right graph is the zoomed-in region with high prediction specificities (0.9–1).
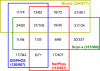
Fig. 7.
Prediction consistency among different tools at specificity around 95% on same test results as in Fig. 6. Different colors indicate different tools. Blocks with edges of different colors represent overlapping predictions from corresponding tools. The numbers in each block represent the number of true positives and the number of predicted phosphorylation sites separated by a slash. The numbers in the parentheses following each tool name have a similar meaning for all the predicted sites by the tool.
Fig. 8.
Screenshot of Musite V1. 0 graphical user interface. As an example, the phosphoserine/threonine prediction result of human p53 is displayed.
Similar articles
- The Musite open-source framework for phosphorylation-site prediction.
Gao J, Xu D. Gao J, et al. BMC Bioinformatics. 2010 Dec 21;11 Suppl 12(Suppl 12):S9. doi: 10.1186/1471-2105-11-S12-S9. BMC Bioinformatics. 2010. PMID: 21210988 Free PMC article. - Incorporating key position and amino acid residue features to identify general and species-specific Ubiquitin conjugation sites.
Chen X, Qiu JD, Shi SP, Suo SB, Huang SY, Liang RP. Chen X, et al. Bioinformatics. 2013 Jul 1;29(13):1614-22. doi: 10.1093/bioinformatics/btt196. Epub 2013 Apr 26. Bioinformatics. 2013. PMID: 23626001 - Predicting and analyzing protein phosphorylation sites in plants using musite.
Yao Q, Gao J, Bollinger C, Thelen JJ, Xu D. Yao Q, et al. Front Plant Sci. 2012 Aug 21;3:186. doi: 10.3389/fpls.2012.00186. eCollection 2012. Front Plant Sci. 2012. PMID: 22934099 Free PMC article. - Computational prediction of eukaryotic phosphorylation sites.
Trost B, Kusalik A. Trost B, et al. Bioinformatics. 2011 Nov 1;27(21):2927-35. doi: 10.1093/bioinformatics/btr525. Epub 2011 Sep 16. Bioinformatics. 2011. PMID: 21926126 Review. - Towards more accurate prediction of ubiquitination sites: a comprehensive review of current methods, tools and features.
Chen Z, Zhou Y, Zhang Z, Song J. Chen Z, et al. Brief Bioinform. 2015 Jul;16(4):640-57. doi: 10.1093/bib/bbu031. Epub 2014 Sep 10. Brief Bioinform. 2015. PMID: 25212598 Review.
Cited by
- Unveiling orphan receptor-like kinases in plants: novel client discovery using high-confidence library predictions in the Kinase-Client (KiC) assay.
Jorge GL, Kim D, Xu C, Cho SH, Su L, Xu D, Bartley LE, Stacey G, Thelen JJ. Jorge GL, et al. Front Plant Sci. 2024 Apr 3;15:1372361. doi: 10.3389/fpls.2024.1372361. eCollection 2024. Front Plant Sci. 2024. PMID: 38633461 Free PMC article. - 14-3-3 binding motif phosphorylation disrupts Hdac4-organized condensates to stimulate cardiac reprogramming.
Liu L, Lei I, Tian S, Gao W, Guo Y, Li Z, Sabry Z, Tang P, Chen YE, Wang Z. Liu L, et al. Cell Rep. 2024 Apr 23;43(4):114054. doi: 10.1016/j.celrep.2024.114054. Epub 2024 Apr 4. Cell Rep. 2024. PMID: 38578832 Free PMC article. - DOTAD: A Database of Therapeutic Antibody Developability.
Li W, Lin H, Huang Z, Xie S, Zhou Y, Gong R, Jiang Q, Xiang C, Huang J. Li W, et al. Interdiscip Sci. 2024 Sep;16(3):623-634. doi: 10.1007/s12539-024-00613-2. Epub 2024 Mar 26. Interdiscip Sci. 2024. PMID: 38530613 - Identifying Protein Phosphorylation Site-Disease Associations Based on Multi-Similarity Fusion and Negative Sample Selection by Convolutional Neural Network.
Deng Q, Zhang J, Liu J, Liu Y, Dai Z, Zou X, Li Z. Deng Q, et al. Interdiscip Sci. 2024 Sep;16(3):649-664. doi: 10.1007/s12539-024-00615-0. Epub 2024 Mar 8. Interdiscip Sci. 2024. PMID: 38457108 - Attenphos: General Phosphorylation Site Prediction Model Based on Attention Mechanism.
Song T, Yang Q, Qu P, Qiao L, Wang X. Song T, et al. Int J Mol Sci. 2024 Jan 26;25(3):1526. doi: 10.3390/ijms25031526. Int J Mol Sci. 2024. PMID: 38338804 Free PMC article.
References
- Johnson L. N. (2009) The regulation of protein phosphorylation. Biochem. Soc. Trans. 37, 627–641 - PubMed
- Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 - PubMed
- Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., Hunt D. F. (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2193–2198 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R33 GM078601-05/GM/NIGMS NIH HHS/United States
- R33 GM078601-04/GM/NIGMS NIH HHS/United States
- R21/R33 GM078601/GM/NIGMS NIH HHS/United States
- R21 GM078601/GM/NIGMS NIH HHS/United States
- R33 GM078601-03/GM/NIGMS NIH HHS/United States
- R33 GM078601/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources