Antimalarial drug targets in Plasmodium falciparum predicted by stage-specific metabolic network analysis - PubMed (original) (raw)
Antimalarial drug targets in Plasmodium falciparum predicted by stage-specific metabolic network analysis
Carola Huthmacher et al. BMC Syst Biol. 2010.
Abstract
Background: Despite enormous efforts to combat malaria the disease still afflicts up to half a billion people each year of which more than one million die. Currently no approved vaccine is available and resistances to antimalarials are widely spread. Hence, new antimalarial drugs are urgently needed.
Results: Here, we present a computational analysis of the metabolism of Plasmodium falciparum, the deadliest malaria pathogen. We assembled a compartmentalized metabolic model and predicted life cycle stage specific metabolism with the help of a flux balance approach that integrates gene expression data. Predicted metabolite exchanges between parasite and host were found to be in good accordance with experimental findings when the parasite's metabolic network was embedded into that of its host (erythrocyte). Knock-out simulations identified 307 indispensable metabolic reactions within the parasite. 35 out of 57 experimentally demonstrated essential enzymes were recovered and another 16 enzymes, if additionally the assumption was made that nutrient uptake from the host cell is limited and all reactions catalyzed by the inhibited enzyme are blocked. This predicted set of putative drug targets, shown to be enriched with true targets by a factor of at least 2.75, was further analyzed with respect to homology to human enzymes, functional similarity to therapeutic targets in other organisms and their predicted potency for prophylaxis and disease treatment.
Conclusions: The results suggest that the set of essential enzymes predicted by our flux balance approach represents a promising starting point for further drug development.
Figures
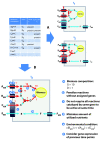
Figure 1
Illustration of presented flux balance approach to predict life cycle specific metabolism. Given the gene expression data (blue table) flux distributions (red arrows) within the shown example metabolic network (blue arrows) can be inferred for time points _t_1 and _t_2 as depicted in (A). However, neither flux direction nor flux strength can be deduced from gene expression alone (indicated by question marks next to flux arrows). The set of all possible flux distributions that are consistent with the gene expression data can be reduced by knowledge about target fluxes such as biomass production (i). Reactions that are not supported by genome annotation might represent errors in the network assembly. Therefore it is desirable to prevent the usage of such reactions in calculated flux distributions (ii). An enzyme or a transporter that is able to process different metabolites does not necessarily convert all substrates at same rates. If one reaction product is not converted further by subsequent enzymes, it accumulates and as a consequence the net production rate is close to zero, even if the gene is expressed and substrate is available (iii). The flux solution space can be narrowed down further when assuming that biomass production is achieved with a minimal amount of nutrients (iv), which are of varying availability (v). Gene products can be present within a cell, even when the gene transcript is not detectable, as proteins appear later than the corresponding mRNA and protein degradation might be delayed compared to mRNA degradation. Considering proteins to be present whose transcript was detectable during a previous time point (vi) presumably reflects the actual cellular status better than taking only the current transcription snapshot into account. The flux distribution calculated by our flux balance approach, which incorporates all these issues, is shown in (B).
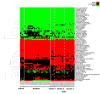
Figure 2
Predicted host parasite metabolite exchanges using improved algorithm. Flux distributions have been predicted with our improved flux balance approach (see Figure 1) for each time point of the intraerythrocytic developmental cycle for which a gene expression profile exists. Simulations were conducted on the basis of the combined metabolic network of parasite and host and additional constraints reflecting knowledge about the blood stage. Furthermore, the expression status of genes during preceding time points was considered for the flux calculations. Resulting metabolite exchanges between host and parasite are depicted in this figure. Red matrix entries represent metabolites that are predicted to be imported into the parasite, while green matrix entries represent metabolites secreted into the host compartment.
Figure 3
Evaluation of predicted drug targets. We conducted FBA based knock-out simulations to uncover reactions within the metabolic network of the parasite that are essential for the production of metabolites assumed to be important for parasite development (see Additional file9). The predicted set of indispensable reactions, which presumably represent good drug targets, was evaluated on the basis of a gold standard set that contains 57 experimentally verified essential enzymes. We determined true positives (TP), false positives (FP), false negatives (FN), and true negatives (TN) and calculated based on these numbers sensitivity, specificity, accuracy and precision of our method as well as the corresponding enrichment factor (red numbers). To compare our method to a previously proposed method for drug target detection, the choke-point analysis, we additionally identified all choke-points within the parasite's metabolic network and calculated the same statistics (green numbers).
Similar articles
- In silico multiple-targets identification for heme detoxification in the human malaria parasite Plasmodium falciparum.
Phaiphinit S, Pattaradilokrat S, Lursinsap C, Plaimas K. Phaiphinit S, et al. Infect Genet Evol. 2016 Jan;37:237-44. doi: 10.1016/j.meegid.2015.11.025. Epub 2015 Dec 2. Infect Genet Evol. 2016. PMID: 26626103 - Network-based assessment of the selectivity of metabolic drug targets in Plasmodium falciparum with respect to human liver metabolism.
Bazzani S, Hoppe A, Holzhütter HG. Bazzani S, et al. BMC Syst Biol. 2012 Aug 31;6:118. doi: 10.1186/1752-0509-6-118. BMC Syst Biol. 2012. PMID: 22937810 Free PMC article. - Novel Plasmodium falciparum metabolic network reconstruction identifies shifts associated with clinical antimalarial resistance.
Carey MA, Papin JA, Guler JL. Carey MA, et al. BMC Genomics. 2017 Jul 19;18(1):543. doi: 10.1186/s12864-017-3905-1. BMC Genomics. 2017. PMID: 28724354 Free PMC article. - A proteomic glimpse into the effect of antimalarial drugs on Plasmodium falciparum proteome towards highlighting possible therapeutic targets.
Dousti M, Manzano-Román R, Rashidi S, Barzegar G, Ahmadpour NB, Mohammadi A, Hatam G. Dousti M, et al. Pathog Dis. 2021 Jan 9;79(1):ftaa071. doi: 10.1093/femspd/ftaa071. Pathog Dis. 2021. PMID: 33202000 Review. - Supply and demand-heme synthesis, salvage and utilization by Apicomplexa.
Kloehn J, Harding CR, Soldati-Favre D. Kloehn J, et al. FEBS J. 2021 Jan;288(2):382-404. doi: 10.1111/febs.15445. Epub 2020 Jun 23. FEBS J. 2021. PMID: 32530125 Review.
Cited by
- A review on computational systems biology of pathogen-host interactions.
Durmuş S, Çakır T, Özgür A, Guthke R. Durmuş S, et al. Front Microbiol. 2015 Apr 9;6:235. doi: 10.3389/fmicb.2015.00235. eCollection 2015. Front Microbiol. 2015. PMID: 25914674 Free PMC article. Review. - Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence.
Lobel L, Sigal N, Borovok I, Ruppin E, Herskovits AA. Lobel L, et al. PLoS Genet. 2012 Sep;8(9):e1002887. doi: 10.1371/journal.pgen.1002887. Epub 2012 Sep 6. PLoS Genet. 2012. PMID: 22969433 Free PMC article. - Metabolic reconstruction identifies strain-specific regulation of virulence in Toxoplasma gondii.
Song C, Chiasson MA, Nursimulu N, Hung SS, Wasmuth J, Grigg ME, Parkinson J. Song C, et al. Mol Syst Biol. 2013 Nov 19;9:708. doi: 10.1038/msb.2013.62. Mol Syst Biol. 2013. PMID: 24247825 Free PMC article. - Promise and reality in the expanding field of network interaction analysis: metabolic networks.
Bazzani S. Bazzani S. Bioinform Biol Insights. 2014 Apr 16;8:83-91. doi: 10.4137/BBI.S12466. eCollection 2014. Bioinform Biol Insights. 2014. PMID: 24812497 Free PMC article. - Antiplasmodial dihetarylthioethers target the coenzyme A synthesis pathway in Plasmodium falciparum erythrocytic stages.
Weidner T, Lucantoni L, Nasereddin A, Preu L, Jones PG, Dzikowski R, Avery VM, Kunick C. Weidner T, et al. Malar J. 2017 May 15;16(1):192. doi: 10.1186/s12936-017-1839-3. Malar J. 2017. PMID: 28502250 Free PMC article.
References
- WHO. World Malaria Report 2008 http://apps.who.int/malaria/wmr2008/malaria 2008.pdf. Tech rep World Health Organization. 2008.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources