Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line - PubMed (original) (raw)
Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line
Keiichi Ohshima et al. PLoS One. 2010.
Abstract
Background: Exosomes play a major role in cell-to-cell communication, targeting cells to transfer exosomal molecules including proteins, mRNAs, and microRNAs (miRNAs) by an endocytosis-like pathway. miRNAs are small noncoding RNA molecules on average 22 nucleotides in length that regulate numerous biological processes including cancer pathogenesis and mediate gene down-regulation by targeting mRNAs to induce RNA degradation and/or interfering with translation. Recent reports imply that miRNAs can be stably detected in circulating plasma and serum since miRNAs are packaged by exosomes to be protected from RNA degradation. Thus, profiling exosomal miRNAs are in need to clarify intercellular signaling and discover a novel disease marker as well.
Methodology/principal findings: Exosomes were isolated from cultured cancer cell lines and their quality was validated by analyses of transmission electron microscopy and western blotting. One of the cell lines tested, a metastatic gastric cancer cell line, AZ-P7a, showed the highest RNA yield in the released exosomes and distinctive shape in morphology. In addition, RNAs were isolated from cells and culture media, and profiles of these three miRNA fractions were obtained using microarray analysis. By comparing signal intensities of microarray data and the following validation using RT-PCR analysis, we found that let-7 miRNA family was abundant in both the intracellular and extracellular fractions from AZ-P7a cells, while low metastatic AZ-521, the parental cell line of AZ-P7a, as well as other cancer cell lines showed no such propensity.
Conclusions/significance: The enrichment of let-7 miRNA family in the extracellular fractions, particularly, in the exosomes from AZ-P7a cells may reflect their oncogenic characteristics including tumorigenesis and metastasis. Since let-7 miRNAs generally play a tumor-suppressive role as targeting oncogenes such as RAS and HMGA2, our results suggest that AZ-P7a cells release let-7 miRNAs via exosomes into the extracellular environment to maintain their oncogenesis.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Characterization of exosomes.
A. Morphological characterization of exosomes derived from AZ-P7a, AZ-521, and SBC-3 cells by transmission electron microscopy. B. Immunoelectron micrographs of AZ-P7a exosomes labeled with immunogold anti-CD63. Exosomes with black colloidal gold particles on the capsular membranes were observed as positive CD 63 staining under the transmission electron microscope (arrowheads). C. Molecular characterization of exosomes derived from AZ-P7a, AZ-521, and SBC-3 cells by western blot analysis. Protein extracts (10 µg) prepared from cells (C) or exosomes (Ex) were assessed using antibodies against exosomal protein markers (CD29/β1-integrin, Aip1/Alix, and Tsg101) and an endoplasmic reticulum marker (Bip/Grp78).
Figure 2. Total RNAs in exosomes and culture media from various cancer cell lines.
A. Amounts of total RNAs recovered from exosomes (lower panel) and culture media (upper panel). The amount of total RNAs per cell was shown. B. Distribution in length of RNA. Isolated total RNAs from the exosomes and culture medium of AZ-P7a cells were detected using Bioanalyzer. The result of cellular total RNAs was shown for comparison.
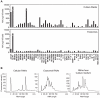
Figure 3. miRNA profiling in the intra- and extra-cellular fractions from AZ-P7a, AZ-521, NCI-H69, SBC-3, DMS53, SW480 and SW620 cells.
miRNA profiles in cells (C), exosomes (Ex), and culture media (CM) were obtained by miRNA microarray analysis. Y axis represents log2 of hybridization signals, shown by bars and filled boxes. The arrows on the left side from the bunch of signals represent signals corresponding to let-7 miRNA family including let-7a (red), let-7b (yellow), let-7c (light green), let-7d (dark green), let-7e (sky blue), let-7f (blue), let-7g (navy blue), and let-7i (purple).
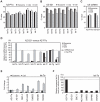
Figure 4. RT-PCR analyses of intra- and extra-cellular let-7 miRNA family.
A, B. Levels of mature let-7 miRNA family including let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, and let-7i in cells (shaded bars), exosomes (filled bars), and culture medium (open bars) from AZ-P7a cells (A) and AZ-521 cells (B). Each miRNA level was analyzed using quantitative RT-PCR. The cycle threshold (Ct) value is presented as the mean ± SD (n = 4). C. Levels of U6 snRNA in cells (shaded bars), exosomes (filled bars), and culture medium (open bars) from AZ-P7a and AZ-521 cells detected by quantitative RT-PCR analysis. The cycle threshold (Ct) value is presented as the mean ± SD (n = 4). D. Relative amounts of let-7 miRNA family in cells (shaded bars), exosomes (filled bars), and culture medium (open bars) from AZ-521 cells versus AZ-P7a cells. The dotted line and arrow represents the levels of let-7 miRNA family in AZ-P7a cells, shown as 100%. The levels of U6 snRNA in each sample were used as an internal standard for normalization amounts of let-7 miRNAs. Except let-7f and let-7g, the levels of let-7 miRNAs were reduced in the extracellular fractions. E. Relative amounts of let-7a in cells (shaded bars) and exosomes (filled bars) from 7 cancer cell lines including AZ-P7a, AZ-521, NCI-H69, SBC-3, DMS53, SW480 and SW620 cells. The dotted line represents the levels in AZ-P7a cells, shown as 1. The levels of U6 snRNA in each sample were used for normalized amounts of let-7a. The value of fold change is presented as the mean ± SD (n = 3) from samples independently prepared from cell culture. F. Relative amounts of exosomal let-7a in 7 cancer cell lines. The dotted line represents the level in AZ-P7a cells, shown as 100%.
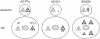
Figure 5. Models on difference in localization of let-7 miRNA family.
Based on the results obtained by microarray and RT-PCR analyses, these models were drawn. In comparison between AZ-P7a cells (left) and AZ-521cells (center), normalized by the amount of U6 snRNA (open triangles), the amount of exosomal let-7a miRNA (filled triangles) in AZ-521 cells was approximately 3% of that in AZ-P7a cells while the intracellular amount in AZ-521 cells was rather 1.4 times greater than that in AZ-P7a cells (Figure 4E). These models are applied to six of eight let-7 miRNA family tested, including let-7a, let-7b, let-7c, let-7d, let-7e, and let-7i. In SW620 cells (right), the amount of let-7a was 3.5 times greater than that in AZ-P7a cells while the amount of exosomal let-7a was 0.7 times less than in AZ-P7a cells (Figure 4E).
Similar articles
- CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway.
Li C, Liu DR, Li GG, Wang HH, Li XW, Zhang W, Wu YL, Chen L. Li C, et al. World J Gastroenterol. 2015 May 28;21(20):6215-28. doi: 10.3748/wjg.v21.i20.6215. World J Gastroenterol. 2015. PMID: 26034356 Free PMC article. - Purification and microRNA profiling of exosomes derived from blood and culture media.
McDonald MK, Capasso KE, Ajit SK. McDonald MK, et al. J Vis Exp. 2013 Jun 14;(76):e50294. doi: 10.3791/50294. J Vis Exp. 2013. PMID: 23792786 Free PMC article. - Exosomal miRNAs from Peritoneum Lavage Fluid as Potential Prognostic Biomarkers of Peritoneal Metastasis in Gastric Cancer.
Tokuhisa M, Ichikawa Y, Kosaka N, Ochiya T, Yashiro M, Hirakawa K, Kosaka T, Makino H, Akiyama H, Kunisaki C, Endo I. Tokuhisa M, et al. PLoS One. 2015 Jul 24;10(7):e0130472. doi: 10.1371/journal.pone.0130472. eCollection 2015. PLoS One. 2015. PMID: 26208314 Free PMC article. - Exosomes-microRNAs interacted with gastric cancer and its microenvironment: a mini literature review.
Deng Z, Wu J, Xu S, Chen F, Zhang Z, Jin A, Wang J. Deng Z, et al. Biomark Med. 2020 Feb;14(2):141-150. doi: 10.2217/bmm-2019-0387. Epub 2020 Feb 17. Biomark Med. 2020. PMID: 32064893 Review. - MicroRNAs in Cancer: From Gene Expression Regulation to the Metastatic Niche Reprogramming.
Semina EV, Rysenkova KD, Troyanovskiy KE, Shmakova AA, Rubina KA. Semina EV, et al. Biochemistry (Mosc). 2021 Jul;86(7):785-799. doi: 10.1134/S0006297921070014. Biochemistry (Mosc). 2021. PMID: 34284705 Review.
Cited by
- Circulating miRNAs as biomarkers for endocrine disorders.
Butz H, Kinga N, Racz K, Patocs A. Butz H, et al. J Endocrinol Invest. 2016 Jan;39(1):1-10. doi: 10.1007/s40618-015-0316-5. Epub 2015 May 28. J Endocrinol Invest. 2016. PMID: 26015318 Review. - Advances in the role of exosomal non-coding RNA in the development, diagnosis, and treatment of gastric cancer (Review).
Gao PF, Huang D, Wen JY, Liu W, Zhang HW. Gao PF, et al. Mol Clin Oncol. 2020 Aug;13(2):101-108. doi: 10.3892/mco.2020.2068. Epub 2020 Jun 10. Mol Clin Oncol. 2020. PMID: 32714531 Free PMC article. Review. - Circulating exosomal miRNAs as potential biomarkers for Barrett's esophagus and esophageal adenocarcinoma.
Lv J, Zhao HP, Dai K, Cheng Y, Zhang J, Guo L. Lv J, et al. World J Gastroenterol. 2020 Jun 14;26(22):2889-2901. doi: 10.3748/wjg.v26.i22.2889. World J Gastroenterol. 2020. PMID: 32587437 Free PMC article. Review. - Extracellular circulating viral microRNAs: current knowledge and perspectives.
Laganà A, Russo F, Veneziano D, Bella SD, Giugno R, Pulvirenti A, Croce CM, Ferro A. Laganà A, et al. Front Genet. 2013 Jun 24;4:120. doi: 10.3389/fgene.2013.00120. eCollection 2013. Front Genet. 2013. PMID: 23805153 Free PMC article. - Horizontal transfer of microRNAs: molecular mechanisms and clinical applications.
Chen X, Liang H, Zhang J, Zen K, Zhang CY. Chen X, et al. Protein Cell. 2012 Jan;3(1):28-37. doi: 10.1007/s13238-012-2003-z. Epub 2012 Feb 9. Protein Cell. 2012. PMID: 22314808 Free PMC article. Review.
References
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. - PubMed
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. - PubMed
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. - PubMed
- Davis DM. Intercellular transfer of cell-surface proteins is common and can affect many stages of an immune response. Nat Rev Immunol. 2007;7:238–243. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases