CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function - PubMed (original) (raw)
. 2010 Nov 2;107(44):19090-5.
doi: 10.1073/pnas.1014523107. Epub 2010 Oct 18.
Xiping Zhang, John J McCarthy, Erin L McDearmon, Troy A Hornberger, Brenda Russell, Kenneth S Campbell, Sandrine Arbogast, Michael B Reid, John R Walker, John B Hogenesch, Joseph S Takahashi, Karyn A Esser
Affiliations
- PMID: 20956306
- PMCID: PMC2973897
- DOI: 10.1073/pnas.1014523107
CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function
Jessica L Andrews et al. Proc Natl Acad Sci U S A. 2010.
Abstract
MyoD, a master regulator of myogenesis, exhibits a circadian rhythm in its mRNA and protein levels, suggesting a possible role in the daily maintenance of muscle phenotype and function. We report that MyoD is a direct target of the circadian transcriptional activators CLOCK and BMAL1, which bind in a rhythmic manner to the core enhancer of the MyoD promoter. Skeletal muscle of Clock(Δ19) and Bmal1(-/-) mutant mice exhibited ∼30% reductions in normalized maximal force. A similar reduction in force was observed at the single-fiber level. Electron microscopy (EM) showed that the myofilament architecture was disrupted in skeletal muscle of Clock(Δ19), Bmal1(-/-), and MyoD(-/-) mice. The alteration in myofilament organization was associated with decreased expression of actin, myosins, titin, and several MyoD target genes. EM analysis also demonstrated that muscle from both Clock(Δ19) and Bmal1(-/-) mice had a 40% reduction in mitochondrial volume. The remaining mitochondria in these mutant mice displayed aberrant morphology and increased uncoupling of respiration. This mitochondrial pathology was not seen in muscle of MyoD(-/-) mice. We suggest that altered expression of both Pgc-1α and Pgc-1β in Clock(Δ19) and Bmal1(-/-) mice may underlie this pathology. Taken together, our results demonstrate that disruption of CLOCK or BMAL1 leads to structural and functional alterations at the cellular level in skeletal muscle. The identification of MyoD as a clock-controlled gene provides a mechanism by which the circadian clock may generate a muscle-specific circadian transcriptome in an adaptive role for the daily maintenance of adult skeletal muscle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
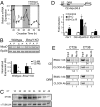
MyoD is a clock-controlled gene in skeletal muscle. (A) Expression of MyoD in wild type (●) and ClockΔ19 (○) muscle was determined by quantitative PCR. Samples were collected every 4 h for 48 h starting at circadian time 18 (CT18) through CT62. Even though all collections were performed under total darkness, the dark and light stripes on the graph represent presumptive dark and light phases of the mice. (B) The diurnal expression (12:00 AM vs. 12:00 PM) of MyoD in wild-type (lanes 1–4) and Bmal1−/− (lanes 5–8) skeletal muscle was determined by semiquantitative PCR normalized to Rpl26 gene expression. Muscles (n = 4/group) were collected under DD at either 12:00 AM (lanes 1, 3, 5, and 7) or 12:00 PM (lanes 2, 4, 6, and 8). Histogram of densitometric quantification showed a significant (P < 0.05) diurnal expression of MyoD in wild-type muscle that is lost in Bmal1−/− muscle. (C) Western blots demonstrating circadian oscillation of MyoD levels in muscle of wild-type mice collected every 4 h for 28 h (CT18–46). (D) Illustration of MyoD reporter gene (CE+_MyoD_6.8) showing the position of the CE and DRR. The histogram summarizes results from transfection experiments using either a Per1 reporter gene or the MyoD reporter gene in C2C12 cells (n = 3/conditions). Over-expression of CLOCK and BMAL1 (black bar) significantly transactivated Per1 and CE-MyoD reporter genes by ∼2.5-fold and 6-fold, respectively, relative to control transfections (open bar). MyoD reporter was not activated by BMAL1:CLOCK, and activation of CE-MyoD reporter was significantly decreased by 50% when ClockΔ19 was over-expressed with BMAL1 (gray bar). Values are mean ± SEM with significance (P < 0.05) denoted by an asterisk or a pound sign (B+C vs. B+C_Δ_19). (E) Chromatin immunoprecipitation assays from muscles collected at CT26 and CT38 demonstrating CLOCK and BMAL1 binding to the CE at CT38 and no binding at the DRR of the MyoD promoter. The numbers under each lane represent the ratio of the intensity of the Ab band/No Ab band.
Fig. 2.
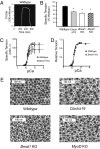
Decreased whole-muscle function, single-cell function, and myofilament structure in ClockΔ19, Bmal−/−, and MyoD−/− mice. (A) Representative force trace from the measurement of specific tension of whole-muscle (EDL) from wild-type mice. (B) Histogram of the average specific tensions of muscles for ClockΔ19, Bmal1−/−, and MyoD−/− mice (n = 3–6/strain). Significant difference (P < 0.05) from wild type is denoted by an asterisk. (C) Results from single-fiber mechanical analyses of wild-type (△) and Bmal1−/− (○) muscle fibers. Each point on the curve represents the average ± SEM for measures of 7–20 cells at each calcium concentration. (D) Data from C reported as tension relative to maximal tension for each calcium concentration. (E) Representative myofilament images obtained by electron microscopy (43,000×) from wild-type, ClockΔ19, Bmal1−/−, and MyoD−/− gastrocnemius muscles. The normal organization of thin and thick filaments is disrupted in muscle from the three different mutant animals.
Fig. 3.
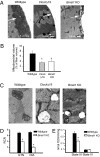
Decreased mitochondrial volume and respiratory function in muscle of ClockΔ19 and Bmal1−/− mice. (A) Low-magnification EM images (4,000×) of skeletal muscle from wild-type, ClockΔ19, and Bmal1−/− mice. The white arrow in each image points to the region of the muscle under the sarcolemma where there are abundant mitochondria (wild type) or where mitochondria are lacking (ClockΔ19 and Bmal1−/−). (B) Histogram of mitochondrial volume measured using point-counting morphometry. The values are presented as a percentage of muscle-fiber volume from wild-type (black bar), ClockΔ19 (gray bar), and Bmal1−/− (open bar) mice. Values represent mean ± SEM (n = 5 muscles/strain) with significance (P < 0.05) denoted by an asterisk. (C) Representative high-magnification EM images (21,000×) of mitochondria within skeletal muscle of wild-type, ClockΔ19, and Bmal1−/− mice. Note swollen size and disrupted cristae of the mitochondria from muscle of ClockΔ19 and Bmal1−/− mice. (D) Histograms of biochemical measurements of respiratory control ratio (RCR) in gastrocnemius (GTN) and diaphragm (DIA) muscles of wild-type and Bmal1−/− mice (n = 6/strain). Values are means ± SEM with significance (P < 0.05) denoted by an asterisk. (E) Histograms showing significant reduction in state III respiration (ADP-stimulated, mmol O2/min/mg protein) in mitochondria isolated from GTN muscle of Bmal1−/− mice compared with wild type. Values are means ± SEM with significance (P < 0.05) indicated by an asterisk.
Fig. 4.
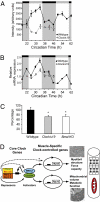
Altered expression of Pgc-1 coactivators in ClockΔ19 and Bmal1−/− mice. (A) Array data of Pgc-1β mRNA expression in skeletal muscle of wild-type mice (●) and ClockΔ19 mice (○); the light and dark stripes refer to the presumptive light and dark phases for the mice (7). (B) Quantitative PCR results for expression of Pgc-1β in wild-type muscle (●) and ClockΔ19 muscle (○). (C) Histogram of the mean expression level of PGC1α mRNA in muscle of wild-type, ClockΔ19, and Bmal1−/− mice as determined by quantitative PCR. A significant difference (P < 0.05) is denoted by an asterisk. (D) Proposed model of CLOCK:BMAL1 regulation of muscle phenotype and function via targeting of MyoD and Pgc-1 expression. Solid lines indicate known molecular links among components of the molecular clock, and dashed lines suggest potential links.
Similar articles
- A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle.
Zhang X, Patel SP, McCarthy JJ, Rabchevsky AG, Goldhamer DJ, Esser KA. Zhang X, et al. Nucleic Acids Res. 2012 Apr;40(8):3419-30. doi: 10.1093/nar/gkr1297. Epub 2011 Dec 30. Nucleic Acids Res. 2012. PMID: 22210883 Free PMC article. - Age-associated disruption of molecular clock expression in skeletal muscle of the spontaneously hypertensive rat.
Miyazaki M, Schroder E, Edelmann SE, Hughes ME, Kornacker K, Balke CW, Esser KA. Miyazaki M, et al. PLoS One. 2011;6(11):e27168. doi: 10.1371/journal.pone.0027168. Epub 2011 Nov 4. PLoS One. 2011. PMID: 22076133 Free PMC article. - Endurance training ameliorates the metabolic and performance characteristics of circadian Clock mutant mice.
Pastore S, Hood DA. Pastore S, et al. J Appl Physiol (1985). 2013 Apr;114(8):1076-84. doi: 10.1152/japplphysiol.01505.2012. Epub 2013 Feb 21. J Appl Physiol (1985). 2013. PMID: 23429867 - The Circadian Clock, Nutritional Signals and Reproduction: A Close Relationship.
Ono M, Ando H, Daikoku T, Fujiwara T, Mieda M, Mizumoto Y, Iizuka T, Kagami K, Hosono T, Nomura S, Toyoda N, Sekizuka-Kagami N, Maida Y, Kuji N, Nishi H, Fujiwara H. Ono M, et al. Int J Mol Sci. 2023 Jan 12;24(2):1545. doi: 10.3390/ijms24021545. Int J Mol Sci. 2023. PMID: 36675058 Free PMC article. Review. - Effects of BMAL1 Manipulation on the Brain's Master Circadian Clock and Behavior.
Haque SN, Booreddy SR, Welsh DK. Haque SN, et al. Yale J Biol Med. 2019 Jun 27;92(2):251-258. eCollection 2019 Jun. Yale J Biol Med. 2019. PMID: 31249486 Free PMC article. Review.
Cited by
- The effect of voluntary exercise on light cycle stress-induced metabolic resistance.
Moon HY, Jeong IC. Moon HY, et al. Phys Act Nutr. 2023 Sep;27(3):1-9. doi: 10.20463/pan.2023.0022. Epub 2023 Sep 30. Phys Act Nutr. 2023. PMID: 37946440 Free PMC article. - BMAL1 drives muscle repair through control of hypoxic NAD+ regeneration in satellite cells.
Zhu P, Hamlish NX, Thakkar AV, Steffeck AWT, Rendleman EJ, Khan NH, Waldeck NJ, DeVilbiss AW, Martin-Sandoval MS, Mathews TP, Chandel NS, Peek CB. Zhu P, et al. Genes Dev. 2022 Feb 1;36(3-4):149-166. doi: 10.1101/gad.349066.121. Epub 2022 Feb 3. Genes Dev. 2022. PMID: 35115380 Free PMC article. - Deletion of Bmal1 Prevents Diet-Induced Ectopic Fat Accumulation by Controlling Oxidative Capacity in the Skeletal Muscle.
Wada T, Ichihashi Y, Suzuki E, Kosuge Y, Ishige K, Uchiyama T, Makishima M, Nakao R, Oishi K, Shimba S. Wada T, et al. Int J Mol Sci. 2018 Sep 18;19(9):2813. doi: 10.3390/ijms19092813. Int J Mol Sci. 2018. PMID: 30231537 Free PMC article. - Circadian misalignment induces fatty acid metabolism gene profiles and compromises insulin sensitivity in human skeletal muscle.
Wefers J, van Moorsel D, Hansen J, Connell NJ, Havekes B, Hoeks J, van Marken Lichtenbelt WD, Duez H, Phielix E, Kalsbeek A, Boekschoten MV, Hooiveld GJ, Hesselink MKC, Kersten S, Staels B, Scheer FAJL, Schrauwen P. Wefers J, et al. Proc Natl Acad Sci U S A. 2018 Jul 24;115(30):7789-7794. doi: 10.1073/pnas.1722295115. Epub 2018 Jul 9. Proc Natl Acad Sci U S A. 2018. PMID: 29987027 Free PMC article. - Deep RNA profiling identified CLOCK and molecular clock genes as pathophysiological signatures in collagen VI myopathy.
Scotton C, Bovolenta M, Schwartz E, Falzarano MS, Martoni E, Passarelli C, Armaroli A, Osman H, Rodolico C, Messina S, Pegoraro E, D'Amico A, Bertini E, Gualandi F, Neri M, Selvatici R, Boffi P, Maioli MA, Lochmüller H, Straub V, Bushby K, Castrignanò T, Pesole G, Sabatelli P, Merlini L, Braghetta P, Bonaldo P, Bernardi P, Foley R, Cirak S, Zaharieva I, Muntoni F, Capitanio D, Gelfi C, Kotelnikova E, Yuryev A, Lebowitz M, Zhang X, Hodge BA, Esser KA, Ferlini A. Scotton C, et al. J Cell Sci. 2016 Apr 15;129(8):1671-84. doi: 10.1242/jcs.175927. Epub 2016 Mar 4. J Cell Sci. 2016. PMID: 26945058 Free PMC article.
References
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–1486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 MH61915/MH/NIMH NIH HHS/United States
- R01 HL045721/HL/NHLBI NIH HHS/United States
- AR053641/AR/NIAMS NIH HHS/United States
- R21 AR050717/AR/NIAMS NIH HHS/United States
- U01 MH061915/MH/NIMH NIH HHS/United States
- R03 AR053641/AR/NIAMS NIH HHS/United States
- R01 AR055246/AR/NIAMS NIH HHS/United States
- HL45721/HL/NHLBI NIH HHS/United States
- AR050717/AR/NIAMS NIH HHS/United States
- P50 MH074924/MH/NIMH NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases