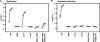The acidic domain of hepatitis C virus NS4A contributes to RNA replication and virus particle assembly - PubMed (original) (raw)
The acidic domain of hepatitis C virus NS4A contributes to RNA replication and virus particle assembly
Tung Phan et al. J Virol. 2011 Feb.
Abstract
Hepatitis C virus NS3-4A is a membrane-bound enzyme complex that exhibits serine protease, RNA helicase, and RNA-stimulated ATPase activities. This enzyme complex is essential for viral genome replication and has been recently implicated in virus particle assembly. To help clarify the role of NS4A in these processes, we conducted alanine scanning mutagenesis on the C-terminal acidic domain of NS4A in the context of a chimeric genotype 2a reporter virus. Of 13 mutants tested, two (Y45A and F48A) had severe defects in replication, while seven (K41A, L44A, D49A, E50A, M51A, E52A, and E53A) efficiently replicated but had severe defects in virus particle assembly. Multiple strategies were used to identify second-site mutations that suppressed these NS4A defects. The replication defect of NS4A F48A was partially suppressed by mutation of NS4B I7F, indicating that a genetic interaction between NS4A and NS4B contributes to RNA replication. Furthermore, the virus assembly defect of NS4A K41A was suppressed by NS3 Q221L, a mutation previously implicated in overcoming other virus assembly defects. We therefore examined the known enzymatic activities of wild-type or mutant forms of NS3-4A but did not detect specific defects in the mutants. Taken together, our data reveal interactions between NS4A and NS4B that control genome replication and between NS3 and NS4A that control virus assembly.
Figures
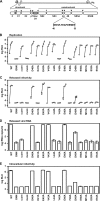
FIG. 1.
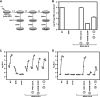
Phenotypes of NS4A mutants. (A) Jc1/GLuc2A reporter construct used in the present study. The sequence of the NS4A C-terminal acidic domain is shown in single-letter amino acid code. Closed circles represent signal peptidase cleavages, the open circle represents a signal peptide peptidase cleavage, the open arrowhead represents NS2-3 cysteine autoprotease cleavage, and closed arrowheads represent NS3-4A serine protease cleavage sites. (B) Replication of NS4A mutants. Cells were transfected with each indicated mutant, and media were collected at 24, 48, 72, and 96 h posttransfection. Values represent the time course of secreted GLuc activity for each transfection, expressed as an average of at least three independent transfections. Error bars represent the standard deviation of the mean. (C) Released infectivity of NS4A mutants. Relative infectivity levels in the media from panel B were quantified by infecting naive cells and monitoring secreted GLuc activity. (D) RNA release of NS4A mutants. The amount of HCV RNA present in the media at 72 h posttransfection was determined by quantitative RT-PCR as described in Materials and Methods. Values represent average RNA quantities from at least three independent transfections. Error bars represent the standard deviation of the mean. (E) Intracellular infectivity of NS4A mutants. Cells were lysed at 48 h posttransfection and used to infect naive cells as described in Materials and Methods. Values represent average relative infectivity measurements from at least three independent transfections. Error bars represent the standard deviation of the mean.
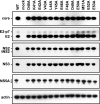
FIG. 2.
Polyprotein processing of NS4A mutants. Parallel cultures of Huh-7.5 cells were infected with vaccinia virus vTF7-3 for 1 h, transfected with the indicated Jc1/GLuc2A cDNA clones, and lysed at 24 h postinfection. Proteins were separated by SDS-PAGE and detected by Western blotting as described in Materials and Methods.
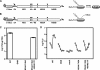
FIG. 3.
NS4B I7F partially suppresses the replication defect of subgenomic replicons containing the NS4A F48A mutation. (A) The pYSGR-JFH construct used in the present study and the workflow to identify revertants or suppressor mutants. HCV polyprotein processing sites are annotated as in Fig. 1. (B) pYSGR-JFH/GLuc reporter construct used in this study and workflow of our reporter virus assay. RNA was electroporated into Huh-7.5 cells, and culture medium was harvested at various times posttransfection. Secreted GLuc activity was assayed as detailed in Materials and Methods. (C) Stable replication phenotypes of NS4A F48A and its suppressor NS4B I7F. Colony-forming activity was measured for NS4A F48A and the suppressor NS4B I7F in the context of pYSGR-JFH. Values represent CFU calculated from three independent transfections. Error bars represent the standard deviation from the mean. (D) Transient replication phenotypes of NS4A F48A and its suppressor NS4B I7F. Luciferase activity was measured for NS4A F48A and the suppressor NS4B I7F in the context of pYSGR-JFH/GLuc. Huh 7.5 cells were transfected with the indicated mutants, and media were collected at 6, 12, 24, 48, 72, and 96 h posttransfection. Values represent the time course of secreted GLuc activity for each transfection, expressed as an average of at least three independent transfections. Error bars represent the standard deviation from the mean.
FIG. 4.
NS4B I7F partially suppresses the replication defect of infectious genomes containing the NS4A F48A mutation. (A) Replication phenotypes of NS4A F48A and its suppressor NS4B I7F. Huh7.5 cells were transfected with each indicated mutant, and medium was collected at 24, 48, 72, and 96 h posttransfection. Values represent the time course of secreted GLuc activity for each transfection, expressed as an average of at least three independent transfections. Error bars represent the standard deviation of the mean. (B) Infectivity phenotypes of NS4A F48A and its suppressor NS4B I7F. The infectivity levels in the medium from panel A were quantified by infecting naive cells and monitoring secreted GLuc activity.
FIG. 5.
NS3 Q221L suppresses the assembly defect of subgenomic replicons containing the NS4A K41A mutation. (A) Workflow used to identify revertants or suppressor mutants via _trans_-packaging. RNAs were electroporated into Huh-7.5[core-NS2] cells, and the cell culture media were collected at each cell passage. The media were used to infect naive cells and productively infected cells were selected with G418. (B) _trans_-Packaging infectivity phenotypes of NS4A K41A and variants containing NS3 Q221L or NS4B S230P. Colony-forming activity was measured for NS4A K41A and the suppressors NS3 Q221L and NS4B S230P in the context of pYSGR-JFH. Values represent the CFU calculated from three independent transfections. Error bars represent the standard deviation from the mean. (C) Replication phenotypes of NS4A K41A and variants containing NS3 Q221L or NS4B S230P. The luciferase activity was measured for NS4A K41A and the suppressor NS3 Q221L and NS4B S230P in the context of pYSGR-JFH/GLuc. Huh-7.5[core-NS2] cells were electroporated with each indicated mutant genome and media were collected at 6, 12, 24, 48, 72, and 96 h posttransfection. Values represent the time course of secreted GLuc activity for each transfection, expressed as an average of at least three independent transfections. Error bars represent the standard deviation of the mean. (D) _trans_-Packaging infectivity phenotypes of NS4A K41A and variants containing NS3 Q221L or NS4B S230P. The infectivity levels in the media from panel C were quantified by infecting naive cells and measuring secreted GLuc activity.
FIG. 6.
NS3 Q221L suppresses the assembly defect of full-length genomes containing the NS4A K41A mutation. (A) Replication phenotypes of NS4A K41A and variants containing NS3 Q221L or NS4B S230P. Huh-7.5 cells were transfected with each indicated mutant, and media were collected at 24, 48, 72, and 96 h posttransfection. Values represent the time course of secreted GLuc activity for each transfection, expressed as an average of at least three independent transfections. Error bars represent the standard deviation of the mean. (B) Relative infectivity phenotypes of NS4A K41A and variants containing NS3 Q221L or NS4B S230P. The levels of infectivity levels in the media from panel A were quantified by infecting naive cells and monitoring secreted GLuc activity. Values represent the time course of secreted GLuc activity for each transfection, expressed as an average of at least three independent transfections. Error bars represent the standard deviation of the mean.
FIG. 7.
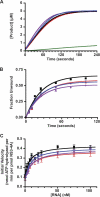
Enzymatic activities of WT and mutant forms of NS3-4A. (A) Serine protease activities of WT (black), NS4A K41A (red), NS3 Q221L (blue), and NS4A K41A + NS3 Q221L (purple). The data were collected at 1-s intervals as described in Materials and Methods. Values represent averages from three independent experiments, and a buffer control (no NS3-4A) is shown in green. (B) RNA unwinding activities of WT (black), NS4A K41A (red), NS3 Q221L (blue), and NS4A K41A + NS3 Q221L (purple) forms of NS3-4A. Values represent the average of three independent experiments. Error bars represent the standard deviation from the mean. (C) RNA-stimulated ATPase activities of WT (black), NS4A K41A (red), NS3 Q221L (blue), and NS4A K41A + NS3 Q221L (purple) forms of NS3-4A. Values represent the average of three independent experiments. Error bars represent standard deviation from the mean.
Similar articles
- Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes.
Phan T, Beran RK, Peters C, Lorenz IC, Lindenbach BD. Phan T, et al. J Virol. 2009 Sep;83(17):8379-95. doi: 10.1128/JVI.00891-09. Epub 2009 Jun 10. J Virol. 2009. PMID: 19515772 Free PMC article. - The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication.
Lindenbach BD, Prágai BM, Montserret R, Beran RK, Pyle AM, Penin F, Rice CM. Lindenbach BD, et al. J Virol. 2007 Sep;81(17):8905-18. doi: 10.1128/JVI.00937-07. Epub 2007 Jun 20. J Virol. 2007. PMID: 17581983 Free PMC article. - The acidic domain of the hepatitis C virus NS4A protein is required for viral assembly and envelopment through interactions with the viral E1 glycoprotein.
Roder AE, Vazquez C, Horner SM. Roder AE, et al. PLoS Pathog. 2019 Feb 7;15(2):e1007163. doi: 10.1371/journal.ppat.1007163. eCollection 2019 Feb. PLoS Pathog. 2019. PMID: 30730994 Free PMC article. - Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus.
Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. Morikawa K, et al. J Viral Hepat. 2011 May;18(5):305-15. doi: 10.1111/j.1365-2893.2011.01451.x. Epub 2011 Mar 23. J Viral Hepat. 2011. PMID: 21470343 Review. - The hepatitis C virus NS3 proteinase: structure and function of a zinc-containing serine proteinase.
De Francesco R, Pessi A, Steinkühler C. De Francesco R, et al. Antivir Ther. 1998;3(Suppl 3):99-109. Antivir Ther. 1998. PMID: 10726060 Review.
Cited by
- Packaging defects in pestiviral NS4A can be compensated by mutations in NS2 and NS3.
Fellenberg J, Dubrau D, Isken O, Tautz N. Fellenberg J, et al. J Virol. 2023 Sep 28;97(9):e0057223. doi: 10.1128/jvi.00572-23. Epub 2023 Sep 11. J Virol. 2023. PMID: 37695056 Free PMC article. - Micro-PET imaging of hepatitis C virus NS3/4A protease activity using a protease-activatable retention probe.
Chuang CH, Cheng TL, Chen WC, Huang YJ, Wang HE, Lo YC, Hsieh YC, Lin WW, Hsieh YJ, Ke CC, Huang KC, Lee JC, Huang MY. Chuang CH, et al. Front Microbiol. 2022 Nov 4;13:896588. doi: 10.3389/fmicb.2022.896588. eCollection 2022. Front Microbiol. 2022. PMID: 36406412 Free PMC article. - Characterization of a multipurpose NS3 surface patch coordinating HCV replicase assembly and virion morphogenesis.
Isken O, Pham MT, Schwanke H, Schlotthauer F, Bartenschlager R, Tautz N. Isken O, et al. PLoS Pathog. 2022 Oct 10;18(10):e1010895. doi: 10.1371/journal.ppat.1010895. eCollection 2022 Oct. PLoS Pathog. 2022. PMID: 36215335 Free PMC article. - The In Vivo and In Vitro Architecture of the Hepatitis C Virus RNA Genome Uncovers Functional RNA Secondary and Tertiary Structures.
Wan H, Adams RL, Lindenbach BD, Pyle AM. Wan H, et al. J Virol. 2022 Apr 27;96(8):e0194621. doi: 10.1128/jvi.01946-21. Epub 2022 Mar 30. J Virol. 2022. PMID: 35353000 Free PMC article. - Hepatitis C Viral Replication Complex.
Li HC, Yang CH, Lo SY. Li HC, et al. Viruses. 2021 Mar 22;13(3):520. doi: 10.3390/v13030520. Viruses. 2021. PMID: 33809897 Free PMC article. Review.
References
- Adair, R., et al. 2009. Expression of hepatitis C virus (HCV) structural proteins in trans facilitates encapsidation and transmission of HCV subgenomic RNA. J. Gen. Virol. 90:833-842. - PubMed
- Beran, R. K., M. M. Bruno, H. A. Bowers, E. Jankowsky, and A. M. Pyle. 2006. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J. Mol. Biol. 358:974-982. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI089826/AI/NIAID NIH HHS/United States
- AI087925/AI/NIAID NIH HHS/United States
- R01 AI087925-01/AI/NIAID NIH HHS/United States
- R01 AI089826/AI/NIAID NIH HHS/United States
- R01 AI087925/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous