L3MBTL1 polycomb protein, a candidate tumor suppressor in del(20q12) myeloid disorders, is essential for genome stability - PubMed (original) (raw)
L3MBTL1 polycomb protein, a candidate tumor suppressor in del(20q12) myeloid disorders, is essential for genome stability
Nadia Gurvich et al. Proc Natl Acad Sci U S A. 2010.
Abstract
The l3mbtl1 gene is located on the long arm of chromosome 20 (q12), within a region commonly deleted in several myeloid malignancies. L3MBTL1 is a human homolog of the Drosophila polycomb L(3)MBT tumor suppressor protein and thus a candidate tumor suppressor in del(20q12) myeloid disorders. We used the loss-of-function approach to explore the possible tumor suppressive mechanism of L3MBTL1 and found that depletion of L3MBTL1 from human cells causes replicative stress, DNA breaks, activation of the DNA damage response, and genomic instability. L3MBTL1 interacts with Cdc45, MCM2-7 and PCNA, components of the DNA replication machinery, and is required for normal replication fork progression, suggesting that L3MBTL1 causes DNA damage, at least in part, by perturbing DNA replication. An activated DNA damage response and genomic instability are common features in tumorigenesis and a consequence of overexpression of many oncogenes. We propose that the loss of L3MBTL1 contributes to the development of 20q(-) hematopoietic malignancies by inducing replicative stress, DNA damage, and genomic instability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
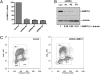
Fig. 1.
L3MBTL1 depletion inhibits cell proliferation and causes G2/M arrest. U2OS cells were infected with control shRNA or one of several different lentiviral shRNAs targeting L3MBTL1. Levels of L3MBTL1 mRNA (A) and protein (B) were measured by quantitative RT-PCR and immunoblotting 48 h after infection. L3MBTL1 mRNA levels detected were adjusted for loading discrepancies using Hprt mRNA as the loading standard, and the levels of mRNA detected were plotted as a percent of the L3MBTL1 observed in U2OS cells infected with control shRNA. To quantitate depletion of L3MBTL1, the relative protein levels of L3MBTL1 were adjusted using the α-tubulin loading control and quantified relative to the protein level present in the control sample (set as 1). (C) U2OS cells were infected with control shRNA or with L3MBTL1 shRNA #3. After 48 h, cells were incubated with BrdU, stained with BrdU-APC antibody and propidium iodide (PI), and analyzed by flow cytometry. The distribution of BrdU (y axis) and PI (x axis) is plotted.
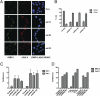
Fig. 2.
Depletion of L3MBTL1 generates DNA breaks. (A) U2OS cells infected with one of several shRNAs against L3MBTL1 or with control shRNA were stained with antibodies against 53BP1 and γH2A.x. (B) DNA damage foci were quantitated in control and L3MBTL1 knockdown cells based on whether they contained more or less than five foci per cell. (C) The tail moment was calculated and plotted from three independent comet assays of U2OS cells treated with control shRNA or L3MBTL1 shRNA U2OS cells at 24 and 48 h after infection. (D) Calculated tail moments from comet assays of control and L3MBTL1-depleted MRC5 and Cal51 cells 48 h after infection are plotted. The tail moment was calculated from comet assays of control MRC5 cells vs. cells irradiated with the indicated dose of gamma irradiation and the data plotted.
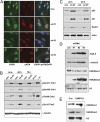
Fig. 3.
Depletion of L3MBTL1 activates the DDR. (A) Control and L3MBTL1-depleted U2OS cells were stained with antibodies against 53BP1 and phospho-ATM (pATM) 48 h postinfection. (B) U2OS cells infected with control or L3MBTL1 shRNAs were harvested 24, 48, and 72 h postinfection and immunoblotted with the indicated antibodies to detect activation of the DDR. Unirradiated cells (unirr) and cells harvested 1 h following 10 Gy of gamma irradiation (γ-irr) were used as negative and positive controls for activated DDR proteins. (C) Lysates from control and L3MBTL1-depleted U2OS cells were isolated 24 and 48 h postinfection, and the levels of Rad51, p53, p21, and actin determined by immunoblotting with the indicated antibodies. (D) Lysates from U2OS cells infected with three different shRNAs against L3MBTL1 were harvested at 48 h and immunoblotted with antibodies directed against γH2A.x, H4K20me1, H4K20me2, histone H3, and tubulin. (E) Histones were extracted from U937 cells that overexpress L3MBTL1 or that contain empty vector control and immunoblotted for H4K20me1, H4K20me2, and histone H3 (loading control).
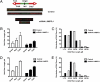
Fig. 4.
Depletion of L3MBTL1 alters the progression of DNA replication forks. MRC5 and U2OS cells infected with control or L3MBTL1 shRNA were incubated for 1 h with IdU followed by 1 h incubation with CldU and then subjected to analysis of replication fork movement. (A) Individual replication units were visualized by immunofluorescence for incorporated halogenated nucleotides in isolated DNA fibers, as described in Materials and Methods. Images of fibers from MRC5 cells infected with control or L3MBTL1 shRNAs are shown. (B) The mean DNA fiber length from MRC5 cells infected with control or L3MBTL1 shRNA was calculated by measuring at least 100 fibers in each experiment, and the results plotted. (C) The data for one representative experiment with MRC5 cells are plotted as percentage of DNA fibers with each specified length. (D) Mean DNA fiber length for U2OS cells infected with control or L3MBTL1 shRNA was calculated by measuring at least 100 fibers in each experiment and plotted. (E) The data derived from an experiment using U2OS cells are plotted as percentage of replication forks with the specified DNA fiber length indicated.
Fig. 5.
L3MBTL1 interacts with components of the DNA replication machinery. (A) HA-L3MBTL1 was overexpressed in 293T cells, and cell lysates were immunoprecipitated and immunoblotted with the HA or rabbit IgG antibodies. (B) Flag-L3MBTL1 was overexpressed in 293T cells, and cell lysates were immunoprecipitated with antibodies against Mcm2, Mcm5, or rabbit IgG. Following SDS/PAGE separation, gels were immunoblotted with Flag antibody to detect L3MBTL1. The immunoprecipitation of Mcm2 and Mcm5 proteins was verified by immunoblotting with the corresponding antibodies, using rabbit IgG as a control.
Similar articles
- Depletion of L3MBTL1 promotes the erythroid differentiation of human hematopoietic progenitor cells: possible role in 20q- polycythemia vera.
Perna F, Gurvich N, Hoya-Arias R, Abdel-Wahab O, Levine RL, Asai T, Voza F, Menendez S, Wang L, Liu F, Zhao X, Nimer SD. Perna F, et al. Blood. 2010 Oct 14;116(15):2812-21. doi: 10.1182/blood-2010-02-270611. Epub 2010 Jun 28. Blood. 2010. PMID: 20585043 Free PMC article. - The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression.
West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E, Kutateladze TG, Gozani O. West LE, et al. J Biol Chem. 2010 Nov 26;285(48):37725-32. doi: 10.1074/jbc.M110.139527. Epub 2010 Sep 24. J Biol Chem. 2010. PMID: 20870725 Free PMC article. - Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1.
Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, Rice JC, Nimer SD. Kalakonda N, et al. Oncogene. 2008 Jul 17;27(31):4293-304. doi: 10.1038/onc.2008.67. Epub 2008 Apr 14. Oncogene. 2008. PMID: 18408754 Free PMC article. - Gene methylation in gastric cancer.
Qu Y, Dang S, Hou P. Qu Y, et al. Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review. - Polycomb Repressor Complex 2 in Genomic Instability and Cancer.
Veneti Z, Gkouskou KK, Eliopoulos AG. Veneti Z, et al. Int J Mol Sci. 2017 Jul 30;18(8):1657. doi: 10.3390/ijms18081657. Int J Mol Sci. 2017. PMID: 28758948 Free PMC article. Review.
Cited by
- Melatonin and Hippo Pathway: Is There Existing Cross-Talk?
Lo Sardo F, Muti P, Blandino G, Strano S. Lo Sardo F, et al. Int J Mol Sci. 2017 Sep 6;18(9):1913. doi: 10.3390/ijms18091913. Int J Mol Sci. 2017. PMID: 28878191 Free PMC article. - Activity-Induced Regulation of Synaptic Strength through the Chromatin Reader L3mbtl1.
Mao W, Salzberg AC, Uchigashima M, Hasegawa Y, Hock H, Watanabe M, Akbarian S, Kawasawa YI, Futai K. Mao W, et al. Cell Rep. 2018 Jun 12;23(11):3209-3222. doi: 10.1016/j.celrep.2018.05.028. Cell Rep. 2018. PMID: 29898393 Free PMC article. - A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation.
Moore KE, Carlson SM, Camp ND, Cheung P, James RG, Chua KF, Wolf-Yadlin A, Gozani O. Moore KE, et al. Mol Cell. 2013 May 9;50(3):444-56. doi: 10.1016/j.molcel.2013.03.005. Epub 2013 Apr 11. Mol Cell. 2013. PMID: 23583077 Free PMC article. - Small-molecule ligands of methyl-lysine binding proteins.
Herold JM, Wigle TJ, Norris JL, Lam R, Korboukh VK, Gao C, Ingerman LA, Kireev DB, Senisterra G, Vedadi M, Tripathy A, Brown PJ, Arrowsmith CH, Jin J, Janzen WP, Frye SV. Herold JM, et al. J Med Chem. 2011 Apr 14;54(7):2504-11. doi: 10.1021/jm200045v. Epub 2011 Mar 18. J Med Chem. 2011. PMID: 21417280 Free PMC article. - Modeling tumor invasion and metastasis in Drosophila.
Miles WO, Dyson NJ, Walker JA. Miles WO, et al. Dis Model Mech. 2011 Nov;4(6):753-61. doi: 10.1242/dmm.006908. Epub 2011 Oct 6. Dis Model Mech. 2011. PMID: 21979943 Free PMC article. Review.
References
- Dewald GW, Schad CR, Lilla VC, Jalal SM. Frequency and photographs of HGM11 chromosome anomalies in bone marrow samples from 3,996 patients with malignant hematologic neoplasms. Cancer Genet Cytogenet. 1993;68:60–69. - PubMed
- Gateff E, Löffler T, Wismar J. A temperature-sensitive brain tumor suppressor mutation of Drosophila melanogaster: Developmental studies and molecular localization of the gene. Mech Dev. 1993;41:15–31. - PubMed
- Min J, et al. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14:1229–1230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous