Deregulation of FoxM1b leads to tumour metastasis - PubMed (original) (raw)
doi: 10.1002/emmm.201000107. Epub 2010 Dec 17.
Galina Gusarova, Zebin Wang, Janai R Carr, Jing Li, Ki-Hyun Kim, Jin Qiu, Yoon-Dong Park, Peter R Williamson, Nissim Hay, Angela L Tyner, Lester F Lau, Robert H Costa, Pradip Raychaudhuri
Affiliations
- PMID: 21204266
- PMCID: PMC3401999
- DOI: 10.1002/emmm.201000107
Deregulation of FoxM1b leads to tumour metastasis
Hyun Jung Park et al. EMBO Mol Med. 2011 Jan.
Abstract
The forkhead box M1b (FoxM1b) transcription factor is over-expressed in human cancers, and its expression often correlates with poor prognosis. Previously, using conditional knockout strains, we showed that FoxM1b is essential for hepatocellular carcinoma (HCC) development. However, over-expression of FoxM1b had only marginal effects on HCC progression. Here we investigated the effect of FoxM1b expression in the absence of its inhibitor Arf. We show that transgenic expression of FoxM1b in an Arf-null background drives hepatic fibrosis and metastasis of HCC. We identify novel mechanisms of FoxM1b that are involved in epithelial-mesenchymal transition, cell motility, invasion and a pre-metastatic niche formation. FoxM1b activates the Akt-Snail1 pathway and stimulates expression of Stathmin, lysyl oxidase, lysyl oxidase like-2 and several other genes involved in metastasis. Furthermore, we show that an Arf-derived peptide, which inhibits FoxM1b, impedes metastasis of the FoxM1b-expressing HCC cells. The observations indicate that FoxM1b is a potent activator of tumour metastasis and that the Arf-mediated inhibition of FoxM1b is a critical mechanism for suppression of tumour metastasis.
Copyright © 2011 EMBO Molecular Medicine.
Figures
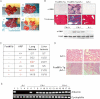
Figure 1. FoxM1b Tg;Arf−/− mice developed fibrosis and metastases of HCC
- We sacrificed the mice and harvested liver tissues after 33 weeks of DEN/PB exposure. FoxM1b Tg;Arf−/− liver tumours (a–c) and lung lesions (d) are shown.
- Liver sections were stained for the activated HSC marker α-SMA or Masson's trichrome for collagen deposition. Magnification: ×200. Lower panel: Liver extracts were subjected to Western blotting assay using α-SMA antibody.
- Numbers of mice bearing lung lesions and liver tumours are shown. For all panels of (B) and (D), the scale bar (indicated in the upper left panel) = 200 µm.
- Lung lesions were stained with hematoxylin and eosin (H&E). Magnification: ×100.
- Total RNA from the lung was extracted for RT-PCR using primers specific for albumin. 1. liver; 2. lung; 3–4. Arf−/− (A: lung); 5–13. FoxM1b Tg;Arf−/− (FA−/−: lung); 14. FoxM1b Tg;Arf−/− (FA−/−: spleen); 15–18. FoxM1b Tg;Arf+/− (FA+/−: lung).
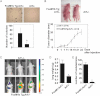
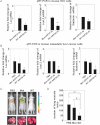
Figure 2. Cells from FoxM1b Tg;Arf−/− HCC are highly tumourigenic and metastatic
- A. Soft agar assays were performed to compare anchorage-independent growth ability. Bar graph presents quantification of the representative data of three independent experiments. Data are expressed as mean ± SD (*p < 0.05).
- B. 5 × 105 cells were subcutaneously injected into nude mice. The tumour diameter was measured at the indicated time points. Data are expressed as mean ± SD.
- C–E. 106 RFP labelled cells were injected into 8-week-old male nude mice intravenously via tail vein. (C) Mice were subjected to fluorescence imaging. (D and E) Mice were then sacrificed and lung tissues were harvested. Lungs were weighed (D) and fixed with Bouin's solution for 24 h. Macroscopic surface tumour nodules were counted (E). Data are expressed as mean ± SD (*p < 0.05).
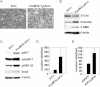
Figure 3. FoxM1b Tg;Arf−/− cells exhibit morphological changes reminiscent of EMT
- Representative phase-contrast images of the cells are shown. Magnification: ×200.
- Expression of EMT markers was determined by immunoblotting. β-Actin served as a loading control.
- Activation of AKT/GSK-3β/Snail pathway was assessed by immunoblotting.
- Invasion assays were performed as described in the Materials and Methods Section.
- Arf−/− HCC cells were infected with adenovirus-expressing GFP or FoxM1b for 48 h and subjected to invasion assay. Data are expressed as mean ± SD (*p < 0.05).
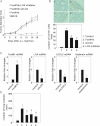
Figure 4. FoxM1b Tg;Arf−/− HCCs express LOX at higher levels compared to Arf−/− HCCs
- A. Semi-quantitative RT-PCR was performed using total RNA from liver tumours.
- B-E. Quantitative RT-PCR was performed to determine mRNA levels of LOX and LOXL2.
- D,E. Arf−/− HCC cells were infected with adenovirus expressing GFP or FoxM1b for 48 h. Data are expressed as mean ± SD (*p < 0.05; **p < 0.01).
- F,G. Chromatin-immunoprecipitation (ChIP) assays were performed using a monoclonal antibody against T7-epitope to detect specific binding of FoxM1b to LOX (F) and LOXL2 (G) promoters in T7-FoxM1b-transfected Sk-hep1 cells.
Figure 5. Infiltration of the Cd11b+ myeloid cells into the lungs of FoxM1b Tg;Arf−/− mice
- A–C. Lung tumour sections were stained for Cd11b+ cells.
- A'–C'. Higher magnification images of the boxed regions in panels A, B and C respectively.
- D, E. Tumour-free lung sections were stained for CdL11b+ cells.
- D'. Higher magnification image of the boxed region in panel D.
- F, G. Lung tumour sections were stained for c-Kit+ cells.
- F'. Higher magnification image of the boxed region in panel F.
- H–J. Collagen accumulation was assessed by Masson's trichrome staining (blue). Scale bar (indicated in panel A) for all lower magnification images = 200 µm.
Figure 6. LOX, LOXL2 and Stathmin play important roles in FoxM1b-mediated metastasis
- A–D. Arf−/− HCC cells were infected with retrovirus expressing FoxM1b and lentivirus expressing shRNA specific to LOX, LOXL2 or Stathmin. Where indicated (in panels A & B) a total of 100 µg/g body weight of a LOX inhibitor, β-aminoproprionitrile (BAPN), was administered daily by i.p. injection starting 1 day prior to injection for the duration of the experiment (18 days).
- A,B. 5 × 105 cells were subcutaneously injected into 8-week-old male nude mice.
- A. The tumour diameter was measured at the indicated time points. Data are expressed as mean ± SD (n = 3).
- B. The mice were sacrificed 21 days after s.c. injection. Lungs were fixed and lung sections were stained with anti-Cd11b antibodies. Upper panels show representative images. Magnification: ×100. Nuclei were stained with hematoxylin (panel 1, control; panel 2, FoxM1b; panel 3, FoxM1b + LOXshRNA; panel 4, FoxM1b + BAPN). Scale bar indicated in panel 1 is same for all panels and equals to 200 µm. Number of Cd11b positive cells per microscopic field was counted (lower panel). A total of 10 fields were randomly chosen. Bar graph presents quantification of the representative data of three independent experiments. Data are expressed as mean ± SD (**p < 0.01 vs. control; #p < 0.05 vs. FoxM1b).
- C. Total RNA was extracted and quantitative RT-PCR was performed to determine mRNA levels of FoxM1, LOX, LOXL2 and Stathmin. Data are expressed as mean ± SD (*p < 0.05; **p < 0.01).
- D. 106 HCC cells were injected into nude mice intravenously via tail vein. Mice were sacrificed and lung tissues were harvested. Lungs were fixed and macroscopic surface tumour nodules were counted. Data are expressed mean ± SD (*p < 0.05 vs. 1; #p < 0.05 vs. 2). 1. MOCK (n = 8); 2. FoxM1b + control (n = 12); 3. FoxM1b + shLOX (n = 5); 4. FoxM1b + shLOX2 (n = 6); 5. FoxM1b + shStathmin (n = 6).
Figure 7. Cell-penetrating ARF peptide inhibitor of FoxM1 prevented pulmonary metastasis of HCC cells
- A,B. FoxM1b Tg;Arf−/− HCC cells (A) or Sk-hep1, a human metastatic cell line (B) were treated with WT ARF26–44 peptide (10 µM) or Mut37–44 peptide (10 µM) for 72 h. Total RNA was extracted and quantitative RT-PCR was performed to determine mRNA levels of LOX, LOXL2 and stathmin. Data are expressed as mean ± SD (*p < 0.05; **p < 0.01).
- C,D. 106 cells labelled with RFP were injected into nude mice intravenously via tail vein. From the next day, mice were i.p. injected with WT ARF peptide, Mut peptide or PBS every other day for 3 weeks at 5 mg/kg body weight. On day 22 from inoculation, mice were subjected to in vivo imaging (C). Mice were then sacrificed and lung tissues were harvested for further quantification. Lungs were fixed with Bouin's solution for 24 h. Macroscopic surface tumour nodules were counted (D). Data are expressed mean ± SD (***p < 0.001; #p < 0.05).
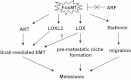
Figure 8. A schematic diagram depicting the various steps of tumour metastasis activated by FoxM1b
FoxM1b activates the Akt pathway and increases expression of LOX and LOXL2 to bring about an EMT-like change. Secreted LOX induces a pre-metastatic niche in the lung. FoxM1-induced expression of Stathmin increases flexibility of the cytoskeleton to enhance cell migration. These mechanisms of FoxM1 contribute to metastasis of HCC.
Similar articles
- FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma.
Meng FD, Wei JC, Qu K, Wang ZX, Wu QF, Tai MH, Liu HC, Zhang RY, Liu C. Meng FD, et al. World J Gastroenterol. 2015 Jan 7;21(1):196-213. doi: 10.3748/wjg.v21.i1.196. World J Gastroenterol. 2015. PMID: 25574092 Free PMC article. - Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor.
Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, Dennewitz MB, Shin B, Datta A, Raychaudhuri P, Costa RH. Kalinichenko VV, et al. Genes Dev. 2004 Apr 1;18(7):830-50. doi: 10.1101/gad.1200704. Genes Dev. 2004. PMID: 15082532 Free PMC article. - [Significance of Forkhead Box m1b (Foxm1b) gene in cell proliferation and carcinogenesis].
Tang SY, Jiao Y, Li LQ. Tang SY, et al. Ai Zheng. 2008 Aug;27(8):894-6. Ai Zheng. 2008. PMID: 18710629 Review. Chinese. - A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment.
Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, Costa RH. Gusarova GA, et al. J Clin Invest. 2007 Jan;117(1):99-111. doi: 10.1172/JCI27527. Epub 2006 Dec 14. J Clin Invest. 2007. PMID: 17173139 Free PMC article. - New and unexpected: forkhead meets ARF.
Costa RH, Kalinichenko VV, Major ML, Raychaudhuri P. Costa RH, et al. Curr Opin Genet Dev. 2005 Feb;15(1):42-8. doi: 10.1016/j.gde.2004.12.007. Curr Opin Genet Dev. 2005. PMID: 15661532 Review.
Cited by
- Targeting FoxM1 effectively retards p53-null lymphoma and sarcoma.
Wang Z, Zheng Y, Park HJ, Li J, Carr JR, Chen YJ, Kiefer MM, Kopanja D, Bagchi S, Tyner AL, Raychaudhuri P. Wang Z, et al. Mol Cancer Ther. 2013 May;12(5):759-67. doi: 10.1158/1535-7163.MCT-12-0903. Epub 2013 Feb 20. Mol Cancer Ther. 2013. PMID: 23427295 Free PMC article. - FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma.
Meng FD, Wei JC, Qu K, Wang ZX, Wu QF, Tai MH, Liu HC, Zhang RY, Liu C. Meng FD, et al. World J Gastroenterol. 2015 Jan 7;21(1):196-213. doi: 10.3748/wjg.v21.i1.196. World J Gastroenterol. 2015. PMID: 25574092 Free PMC article. - Multiple Roles of LOXL2 in the Progression of Hepatocellular Carcinoma and Its Potential for Therapeutic Targeting.
Radić J, Kožik B, Nikolić I, Kolarov-Bjelobrk I, Vasiljević T, Vranjković B, Despotović S. Radić J, et al. Int J Mol Sci. 2023 Jul 21;24(14):11745. doi: 10.3390/ijms241411745. Int J Mol Sci. 2023. PMID: 37511503 Free PMC article. Review. - SUMOylation of FOXM1B alters its transcriptional activity on regulation of MiR-200 family and JNK1 in MCF7 human breast cancer cells.
Wang CM, Liu R, Wang L, Nascimento L, Brennan VC, Yang WH. Wang CM, et al. Int J Mol Sci. 2014 Jun 10;15(6):10233-51. doi: 10.3390/ijms150610233. Int J Mol Sci. 2014. PMID: 24918286 Free PMC article. - FOXM1 predicts overall and disease specific survival in muscle-invasive urothelial carcinoma and presents a differential expression between bladder cancer subtypes.
Rinaldetti S, Wirtz RM, Worst TS, Eckstein M, Weiss CA, Breyer J, Otto W, Bolenz C, Hartmann A, Erben P. Rinaldetti S, et al. Oncotarget. 2017 Jul 18;8(29):47595-47606. doi: 10.18632/oncotarget.17394. Oncotarget. 2017. PMID: 28498805 Free PMC article.
References
- Akiri G, Sabo E, Dafni H, Vadasz Z, Kartvelishvily Y, Gan N, Kessler O, Cohen T, Resnick M, Neeman M, et al. Lysyl oxidase-related protein-1 promotes tumor fibrosis and tumor progression in vivo. Cancer Res. 2003;63:1657–1666. - PubMed
- Baldassarre G, Belletti B, Nicoloso MS, Schiappacassi M, Vecchione A, Spessotto P, Morrione A, Canzonieri V, Colombatti A. p27(Kip1)–stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell. 2005;7:51–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA 124488/CA/NCI NIH HHS/United States
- CA090764/CA/NCI NIH HHS/United States
- R01 AG021842/AG/NIA NIH HHS/United States
- AG025953/AG/NIA NIH HHS/United States
- DK44525/DK/NIDDK NIH HHS/United States
- AG02438/AG/NIA NIH HHS/United States
- AG021842/AG/NIA NIH HHS/United States
- R01 CA090764/CA/NCI NIH HHS/United States
- R01 DK044525/DK/NIDDK NIH HHS/United States
- DK068503/DK/NIDDK NIH HHS/United States
- R01 CA124488/CA/NCI NIH HHS/United States
- R01 GM078492/GM/NIGMS NIH HHS/United States
- R01 CA100035/CA/NCI NIH HHS/United States
- CA 100035/CA/NCI NIH HHS/United States
- R01 DK068503/DK/NIDDK NIH HHS/United States
- R01 AG025953/AG/NIA NIH HHS/United States
- R01 AG016927/AG/NIA NIH HHS/United States
- AG016927/AG/NIA NIH HHS/United States
- I01 BX000131/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous