Proteomics analysis reveals post-translational mechanisms for cold-induced metabolic changes in Arabidopsis - PubMed (original) (raw)
doi: 10.1093/mp/ssq078. Epub 2011 Jan 17.
Shou-Ling Xu, Juan A Oses-Prieto, Sunita Putil, Peng Xu, Rui-Ju Wang, Kathy H Li, David A Maltby, Liz-He An, Alma L Burlingame, Zhi-Ping Deng, Zhi-Yong Wang
Affiliations
- PMID: 21242321
- PMCID: PMC3063518
- DOI: 10.1093/mp/ssq078
Proteomics analysis reveals post-translational mechanisms for cold-induced metabolic changes in Arabidopsis
Tian Li et al. Mol Plant. 2011 Mar.
Abstract
Cold-induced changes of gene expression and metabolism are critical for plants to survive freezing. Largely by changing gene expression, exposure to a period of non-freezing low temperatures increases plant tolerance to freezing-a phenomenon known as cold acclimation. Cold also induces rapid metabolic changes, which provide instant protection before temperature drops below freezing point. The molecular mechanisms for such rapid metabolic responses to cold remain largely unknown. Here, we use two-dimensional difference gel electrophoresis (2-D DIGE) analysis of sub-cellular fractions of Arabidopsis thaliana proteome coupled with spot identification by tandem mass spectrometry to identify early cold-responsive proteins in Arabidopsis. These proteins include four enzymes involved in starch degradation, three HSP100 proteins, several proteins in the tricarboxylic acid cycle, and sucrose metabolism. Upon cold treatment, the Disproportionating Enzyme 2 (DPE2), a cytosolic transglucosidase metabolizing maltose to glucose, increased rapidly in the centrifugation pellet fraction and decreased in the soluble fraction. Consistent with cold-induced inactivation of DPE2 enzymatic activity, the dpe2 mutant showed increased freezing tolerance without affecting the C-repeat binding transcription factor (CBF) transcriptional pathway. These results support a model that cold-induced inactivation of DPE2 leads to rapid accumulation of maltose, which is a cold-induced compatible solute that protects cells from freezing damage. This study provides evidence for a key role of rapid post-translational regulation of carbohydrate metabolic enzymes in plant protection against sudden temperature drop.
Figures
Figure 1.
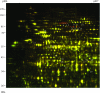
2-D DIGE Analysis of Cold Response in the Soluble Fraction of Arabidopsis Proteome (A) and a MS Spectrum of the PEPC2 Phosphopeptide MASPIDAQLR (B). (A) Seedlings were treated at 2 or 22°C for 2 h, and the soluble protein fractions were compared by 2-D DIGE. In this image, proteins induced by cold treatment appear red, down-regulated appear green, while those unchanged show yellow. Arrows point to cold-responsive spots identified by tandem mass spectrometry. (B) A spectrum of PEPC2 spots (spots 1 and 2 in (A)), corresponding to a phosphopeptide spanning Met-9 to Arg-17. Phosphorylated residue (Ser-11) is indicated in the peptide sequence as SP. The expected and theoretic m/z for the doubly charged peptide was 542.7461 and 542.7465, respectively, and the Error was 0.7 ppm and E-value 0.02.
Figure 2.
2-D DIGE Analysis of Cold Response in the Sodium Carbonate-Soluble Microsomal Fraction of Arabidopsis. Seedlings were treated as in Figure 1, and the microsomal proteins solubilized by 0.1 M sodium carbonate were compared. The cold-induced proteins appear red, and the cold-repressed appear green, while those unchanged show yellow. Arrows point to MS-identified cold-responsive spots.
Figure 3.
2-D DIGE Analysis of Cold Response Proteins in the Triton-Insoluble Fraction. In this image, cold-induced protein spots appear green, and cold-repressed proteins spots appear red.
Figure 4.
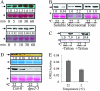
Cold Induces Rapid Accumulation of DPE2 in the Centrifugation Pellet and Decrease of DPE2 Activity in Soluble Cell Extracts. (A) Cold induced a rapid increase of DPE2 in the microsomal (100 000-g pellets) fraction (top panel) but not in the total protein fraction (bottom panel). Arabidopsis seedlings were treated at 2°C for different time duration, and microsomal and total proteins were separated by SDS–PAGE and immunoblotted using the anti-DPE2 antibody. The DPE2 gel bands were quantified using ImageQuant 5.2 software (GE Healthcare) and normalized against the levels of Rubisco large subunit in each sample. Representative images of two repeat experiments performed with different batches of samples are shown. In this figure, the arrows point to DPE2 protein, the stars mark the Ponceau stained band of Rubisco large subunit as the loading control, and the numbers below the gel bands show the normalized signal intensity. (B) DPE2 became less soluble after cold treatment. Seedlings were treated at 2°C for 2 h. Soluble, microsomal, and total proteins were extracted, separated by SDS–PAGE, and immunoblotted using the anti-DPE2 antibody. (C) DPE2 is not directly associated with membranes. Microsome (100 000-g pellets) from both control and cold-treated (2°C for 2 h) samples were re-suspended in microsomal extraction buffer only, or extraction buffer containing 1% (v/v) Triton X-100, and then centrifuged at 100 000 g for 1 h; the pellets were dissolved in SDS buffer and analyzed by anti-DPE2 immunoblot. (D, E) Soluble DPE2 activity was reduced after cold treatment (2°C for 2 h) as shown on native PAGE in-gel assay (D) or assayed in solution (E). (D) Top panel, soluble protein (15 000 g, 10 min) was first separated on native PAGE gel containing 1% oyster glycogen, and then incubated in 5 mM maltose solution, and finally stained with I2-KI solution. The arrow pointed to the band indicates transglucosyl activity, which is absent in the dep2-5 mutant samples. The lower panel shows anti-DPE2 immunoblot of the same protein extracts. (E) DPE2 activity (μmol min−1 g−1 fresh weight) measured in soluble fraction (15 000 g, 10 min) is presented as average of three biological repeats ± standard deviation.
Figure 5.
The dpe2-5 Mutant Is More Tolerant to Freezing Damage. (A) Wild-type (WT) and dpe2 plants were grown at 22°C for 4 weeks, then 4°C for 1 week, and then treated at the indicated temperatures for 1 h and then grown for 1 week at 22°C. (B) Electrolyte leakage in wild-type and dpe2-5 after the 1-h freezing treatments in panel (A). The error bars indicate standard errors (n = 12). (C) Wild-type and dpe2 plants were grown at 22°C for 4 weeks, and then treated at the indicated temperatures for 1 h and then grown for 1 week at 22°C. (D) Electrolyte leakage after 1 h at various temperatures as in (C). The error bars represent standard errors (n = 8). The ion leakage percentage at –6°C was significantly lower in dpe2-5 compared to the wild-type (_p_-value of _t_-test = 0.0034).
Figure 6.
The CBFs and their Downstream Genes Show Similar Kinetics of Response to Cold in Wild-Type and dpe2-5 Plants. Seedlings were treated in ice-cold medium for the indicated time, and expression of individual genes was analyzed by real-time quantitative RT–PCR. The expression of each gene was first normalized to PP2A (At1g13320), and then to the levels of untreated wild-type samples. Data are means ± SD from three biological replicates.
Similar articles
- Temporal proteomics of Arabidopsis plasma membrane during cold- and de-acclimation.
Miki Y, Takahashi D, Kawamura Y, Uemura M. Miki Y, et al. J Proteomics. 2019 Apr 15;197:71-81. doi: 10.1016/j.jprot.2018.11.008. Epub 2018 Nov 14. J Proteomics. 2019. PMID: 30447334 - Starch-related alpha-glucan/water dikinase is involved in the cold-induced development of freezing tolerance in Arabidopsis.
Yano R, Nakamura M, Yoneyama T, Nishida I. Yano R, et al. Plant Physiol. 2005 Jun;138(2):837-46. doi: 10.1104/pp.104.056374. Epub 2005 May 13. Plant Physiol. 2005. PMID: 15894744 Free PMC article. - Cold acclimation by the CBF-COR pathway in a changing climate: Lessons from Arabidopsis thaliana.
Liu Y, Dang P, Liu L, He C. Liu Y, et al. Plant Cell Rep. 2019 May;38(5):511-519. doi: 10.1007/s00299-019-02376-3. Epub 2019 Jan 16. Plant Cell Rep. 2019. PMID: 30652229 Free PMC article. Review. - Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network.
Park S, Lee CM, Doherty CJ, Gilmour SJ, Kim Y, Thomashow MF. Park S, et al. Plant J. 2015 Apr;82(2):193-207. doi: 10.1111/tpj.12796. Epub 2015 Mar 23. Plant J. 2015. PMID: 25736223 - The CBFs: three arabidopsis transcription factors to cold acclimate.
Medina J, Catalá R, Salinas J. Medina J, et al. Plant Sci. 2011 Jan;180(1):3-11. doi: 10.1016/j.plantsci.2010.06.019. Epub 2010 Jul 6. Plant Sci. 2011. PMID: 21421341 Review.
Cited by
- OsDPE2 Regulates Rice Panicle Morphogenesis by Modulating the Content of Starch.
Zheng Y, Fu D, Yang Z. Zheng Y, et al. Rice (N Y). 2023 Feb 3;16(1):5. doi: 10.1186/s12284-023-00618-3. Rice (N Y). 2023. PMID: 36732485 Free PMC article. - Transcriptome Analysis of Low-Temperature-Treated Tetraploid Yellow A ctinidia chinensis Planch. Tissue Culture Plantlets.
Li Y, Zhang Z, Liu X, Wei Z, Zhang X, Bian W, Li S, Zhang H. Li Y, et al. Life (Basel). 2022 Oct 10;12(10):1573. doi: 10.3390/life12101573. Life (Basel). 2022. PMID: 36295009 Free PMC article. - Comparative Transcriptome Analysis Revealing the Potential Mechanism of Low-Temperature Stress in Machilus microcarpa.
He X, Long F, Li Y, Xu Y, Hu L, Yao T, Huang Y, Hu D, Yang Y, Fei Y. He X, et al. Front Plant Sci. 2022 Jul 19;13:900870. doi: 10.3389/fpls.2022.900870. eCollection 2022. Front Plant Sci. 2022. PMID: 35937341 Free PMC article. - Integrative Proteome and Phosphoproteome Profiling of Early Cold Response in Maize Seedlings.
Xing J, Tan J, Feng H, Zhou Z, Deng M, Luo H, Deng Z. Xing J, et al. Int J Mol Sci. 2022 Jun 10;23(12):6493. doi: 10.3390/ijms23126493. Int J Mol Sci. 2022. PMID: 35742945 Free PMC article. - Tandem Mass Tag-Based Quantitative Proteomics Reveals Implication of a Late Embryogenesis Abundant Protein (BnLEA57) in Seed Oil Accumulation in Brassica napus L.
Zhou Z, Lin B, Tan J, Hao P, Hua S, Deng Z. Zhou Z, et al. Front Plant Sci. 2022 Jun 2;13:907244. doi: 10.3389/fpls.2022.907244. eCollection 2022. Front Plant Sci. 2022. PMID: 35720596 Free PMC article.
References
- Amme S, Matros A, Schlesier B, Mock HP. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J. Exp. Bot. 2006;57:1537–1546. - PubMed
- Bae MS, Cho EJ, Choi EY, Park OK. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003;36:652–663. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases