Enteric dysbiosis associated with a mouse model of alcoholic liver disease - PubMed (original) (raw)
Enteric dysbiosis associated with a mouse model of alcoholic liver disease
Arthur W Yan et al. Hepatology. 2011 Jan.
Abstract
The translocation of bacteria and bacterial products into the circulation contributes to alcoholic liver disease. Intestinal bacterial overgrowth is common in patients with alcoholic liver disease. The aims of our study were to investigate bacterial translocation, changes in the enteric microbiome, and its regulation by mucosal antimicrobial proteins in alcoholic liver disease. We used a mouse model of continuous intragastric feeding of alcohol or an isocaloric diet. Bacterial translocation occurred prior to changes observed in the microbiome. Quantitative changes in the intestinal microflora of these animals were assessed first using conventional culture techniques in the small and large intestine. Although we found no difference after 1 day or 1 week, intestinal bacterial overgrowth was observed in the gastrointestinal tract of mice fed alcohol for 3 weeks compared with control mice fed an isocaloric liquid diet. Because <20% of all gastrointestinal bacteria can be cultured using conventional methodologies, we performed massively parallel pyrosequencing to further assess the qualitative changes in the intestinal microbiome following alcohol exposure. Sequencing of 16S ribosomal RNA genes revealed a relative abundance of Bacteroidetes and Verrucomicrobia bacteria in mice fed alcohol compared with a relative predominance of Firmicutes bacteria in control mice. With respect to the host's transcriptome, alcohol feeding was associated with down-regulation in gene and protein expression of bactericidal c-type lectins Reg3b and Reg3g in the small intestine. Treatment with prebiotics partially restored Reg3g protein levels, reduced bacterial overgrowth, and lessened alcoholic steatohepatitis.
Conclusion: Alcohol feeding is associated with intestinal bacterial overgrowth and enteric dysbiosis. Intestinal antimicrobial molecules are dysregulated following chronic alcohol feeding contributing to changes in the enteric microbiome and to alcoholic steatohepatitis.
Copyright © 2010 American Association for the Study of Liver Diseases.
Figures
Figure 1. Intragastric alcohol feeding results in steatohepatitis in mice
Mice were fed alcohol or an isocaloric diet via an intragastric feeding tube. (A) Representative photomicrographs of livers are shown. (B) Plasma ALT levels were measured. (C) Hepatic gene expression of collagen α1(I) was determined by RT-QPCR and normalized to 18S gene expression. Values are presented relative to control fed animals. *p<0.05, when mice fed alcohol are compared to mice fed an isocaloric control diet. Data represent the mean ± SEM of six control or alcohol fed mice at each time point.
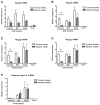
Figure 2. Bacterial translocation to mesenteric lymph nodes or to blood following enteric administration of alcohol
Aerobic bacteria were quantified in (A) the mesenteric lymph nodes and (B) the blood in mice fed with alcohol or an isocaloric diet (n=4–17 animals per time point). *p<0.05 as compared to control fed mice. CFU = colony forming unit. (C) Endotoxin levels were measured and are presented relative to control fed animals for each time point (n=5–10 animals per time point).
Figure 3. Enteric alcohol administration produces intestinal bacterial overgrowth
Total aerobic bacteria (A) and anaerobic bacteria (B) were quantified by culture in the gastrointestinal tract of mice fed alcohol or an isocaloric control diet for three weeks. Data represent the mean ± SEM of twelve control or alcohol fed mice at each time point. *p<0.05, when compared to control treated mice. CFU = colony forming unit. (C) Total bacterial load in the cecum was determined by QPCR using universal bacteria primers. Values are presented relative to control fed animals. *p<0.05, when mice fed alcohol are compared to mice fed an isocaloric control diet. Data represent the mean ± SEM of five control or alcohol fed mice at each time point.
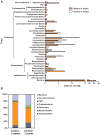
Figure 4. Effects of alcohol on microbial diversity of the mouse cecum
(A) 16S rRNA from the mouse cecum was sequenced using 454 Titanium technology. Experiment-specific OTU representative sequences (97% identity) were classified using the Ribosomal Database Project (RDP) classifier and plotted. Orange bars indicate OTUs containing the alcohol treated group (3 mice, distributed among 349 OTUs) and white bars indicate OTUs containing the isocaloric control group (3 mice, distributed among 297 OTUs). (B) The graph demonstrates the percentages of each community contributed by the indicated phyla. (C) Scatter plots of Unifrac (left) and R (right) PCA. The alcohol treated samples are in blue while the isocaloric control samples are in red.
Figure 4. Effects of alcohol on microbial diversity of the mouse cecum
(A) 16S rRNA from the mouse cecum was sequenced using 454 Titanium technology. Experiment-specific OTU representative sequences (97% identity) were classified using the Ribosomal Database Project (RDP) classifier and plotted. Orange bars indicate OTUs containing the alcohol treated group (3 mice, distributed among 349 OTUs) and white bars indicate OTUs containing the isocaloric control group (3 mice, distributed among 297 OTUs). (B) The graph demonstrates the percentages of each community contributed by the indicated phyla. (C) Scatter plots of Unifrac (left) and R (right) PCA. The alcohol treated samples are in blue while the isocaloric control samples are in red.
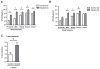
Figure 5. Reg3b and Reg3g gene expression are suppressed by alcohol
Total RNA was prepared from intestinal segments of mice fed with alcohol or isocaloric diet for 1 week (A and C) or 3 weeks (B,D,E). Expression of the Reg3b (A and B), Reg3g (C and D), and Defensin alpha 5 (E) gene were measured by RT-QPCR by using 18S as an internal control. Data represent the mean ± SEM of five control or alcohol fed mice at each time point. *p<0.05, when compared to control treated mice.
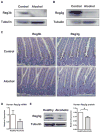
Figure 6. Protein expression of Reg3b and Reg3g is downregulated in the jejunum by alcohol
(A and B) Protein extracts from the proximal small intestine (jejunum) of mice on either an alcohol diet or isocaloric control diet for 3 weeks were analyzed by Western blotting with Reg3b or Reg3g antibodies. Tubulin was used as a loading control. Images are representative of one Western blot, which was reproduced in three independent experiments using different animals each time. (C) Immunohistochemical detection of Reg3b and Reg3g in paraffin-embedded proximal small intestinal sections in mice following alcohol or control feeding for 3 weeks. A representative section is shown. (D) Gene expression of human Reg3g was determined by RT-QPCR and normalized to 18S gene expression in duodenal biopsies from patients with chronic alcohol abuse (n=10) and healthy controls (n=10). Values are presented relative to healthy controls and represent the mean ± SEM. (E) Reg3g and tubulin protein expression in duodenal biopsies from patients with chronic alcohol consumption (n=6) and healthy controls (n=6) were analyzed by Western blot analysis. A representative Western blot image is shown. Densitometry of Western blot images with paired control and alcoholic samples was performed. Values are presented relative to healthy controls and represent the mean ± SEM; *p<0.05.
Figure 7. Prebiotics improve alcoholic steatohepatitis by inducing Reg3g expression and reducing intestinal bacterial overgrowth
Mice were fed an isocaloric diet, alcohol, or alcohol and fructooligosaccharides (FOS) via an intragastric feeding tube for 3 weeks. (A) Reg3g protein expression was analyzed in the proximal small intestine (jejunum) by Western blotting. Tubulin was used as a loading control. Images are representative of one Western blot, which was reproduced in four independent experiments using different animals each time. (B) Total aerobic bacteria (upper graph) and anaerobic bacteria (lower graph) were quantified by culture in the mid small intestine. Data represent the mean ± SEM of 5–6 mice per group; *p<0.05. CFU = colony forming unit. (C) Representative photomicrographs of H&E stained livers are shown. (D) Plasma ALT levels were measured. Data represent the mean ± SEM of 4–14 mice per group; *p<0.05. (E) Hepatic triglyceride content was determined. Data represent the mean ± SEM of 5–10 mice per group; *p<0.05.
Similar articles
- Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice.
Chen P, Torralba M, Tan J, Embree M, Zengler K, Stärkel P, van Pijkeren JP, DePew J, Loomba R, Ho SB, Bajaj JS, Mutlu EA, Keshavarzian A, Tsukamoto H, Nelson KE, Fouts DE, Schnabl B. Chen P, et al. Gastroenterology. 2015 Jan;148(1):203-214.e16. doi: 10.1053/j.gastro.2014.09.014. Epub 2014 Sep 16. Gastroenterology. 2015. PMID: 25239591 Free PMC article. - Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice.
Hartmann P, Chen P, Wang HJ, Wang L, McCole DF, Brandl K, Stärkel P, Belzer C, Hellerbrand C, Tsukamoto H, Ho SB, Schnabl B. Hartmann P, et al. Hepatology. 2013 Jul;58(1):108-19. doi: 10.1002/hep.26321. Epub 2013 May 27. Hepatology. 2013. PMID: 23408358 Free PMC article. - Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice.
Hendrikx T, Duan Y, Wang Y, Oh JH, Alexander LM, Huang W, Stärkel P, Ho SB, Gao B, Fiehn O, Emond P, Sokol H, van Pijkeren JP, Schnabl B. Hendrikx T, et al. Gut. 2019 Aug;68(8):1504-1515. doi: 10.1136/gutjnl-2018-317232. Epub 2018 Nov 17. Gut. 2019. PMID: 30448775 Free PMC article. - Host-microbiome interactions in alcoholic liver disease.
Chen P, Schnabl B. Chen P, et al. Gut Liver. 2014 May;8(3):237-41. doi: 10.5009/gnl.2014.8.3.237. Gut Liver. 2014. PMID: 24827618 Free PMC article. Review. - Alcoholic liver disease: the gut microbiome and liver cross talk.
Hartmann P, Seebauer CT, Schnabl B. Hartmann P, et al. Alcohol Clin Exp Res. 2015 May;39(5):763-75. doi: 10.1111/acer.12704. Alcohol Clin Exp Res. 2015. PMID: 25872593 Free PMC article. Review.
Cited by
- New Developments in Microbiome in Alcohol-Associated and Nonalcoholic Fatty Liver Disease.
Hartmann P, Schnabl B. Hartmann P, et al. Semin Liver Dis. 2021 Jan;41(1):87-102. doi: 10.1055/s-0040-1719174. Epub 2021 Jan 14. Semin Liver Dis. 2021. PMID: 33957682 Free PMC article. - Microbiota reprogramming for treatment of alcohol-related liver disease.
Siddiqui MT, Cresci GAM. Siddiqui MT, et al. Transl Res. 2020 Dec;226:26-38. doi: 10.1016/j.trsl.2020.07.004. Epub 2020 Jul 17. Transl Res. 2020. PMID: 32687975 Free PMC article. Review. - The signatures and crosstalk of gut microbiome, mycobiome, and metabolites in decompensated cirrhotic patients.
Li Y, Liu D, He Y, Zhang Z, Zeng A, Fan C, Lyu L, He Z, Ding H. Li Y, et al. Front Microbiol. 2024 Aug 21;15:1443182. doi: 10.3389/fmicb.2024.1443182. eCollection 2024. Front Microbiol. 2024. PMID: 39234546 Free PMC article. - Gut Microbiome and Alcohol-associated Liver Disease.
Philips CA, Schnabl B, Bajaj JS. Philips CA, et al. J Clin Exp Hepatol. 2022 Sep-Oct;12(5):1349-1359. doi: 10.1016/j.jceh.2021.12.016. Epub 2022 Jan 4. J Clin Exp Hepatol. 2022. PMID: 36157139 Free PMC article. Review. - Toll-like receptors in alcoholic liver disease, non-alcoholic steatohepatitis and carcinogenesis.
Roh YS, Seki E. Roh YS, et al. J Gastroenterol Hepatol. 2013 Aug;28 Suppl 1(0 1):38-42. doi: 10.1111/jgh.12019. J Gastroenterol Hepatol. 2013. PMID: 23855294 Free PMC article. Review.
References
- Bode C, Bode JC. Activation of the innate immune system and alcoholic liver disease: effects of ethanol per se or enhanced intestinal translocation of bacterial toxins induced by ethanol? Alcohol Clin Exp Res. 2005;29:166S–171S. - PubMed
- Brann OS. Infectious complications of cirrhosis. Curr Gastroenterol Rep. 2001;3:285–292. - PubMed
- Christou L, Pappas G, Falagas ME. Bacterial infection-related morbidity and mortality in cirrhosis. Am J Gastroenterol. 2007;102:1510–1517. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K08 DK081830-01/DK/NIDDK NIH HHS/United States
- P50AA11999/AA/NIAAA NIH HHS/United States
- K08 DK081830-02/DK/NIDDK NIH HHS/United States
- K08 DK081830/DK/NIDDK NIH HHS/United States
- R01 DK072237/DK/NIDDK NIH HHS/United States
- P50 AA011999/AA/NIAAA NIH HHS/United States
- R01 GM041804/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources