Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing - PubMed (original) (raw)
Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing
Matthieu Legendre et al. Virol J. 2011.
Abstract
Background: Mimivirus, a giant dsDNA virus infecting Acanthamoeba, is the prototype of the mimiviridae family, the latest addition to the family of the nucleocytoplasmic large DNA viruses (NCLDVs). Its 1.2 Mb-genome was initially predicted to encode 917 genes. A subsequent RNA-Seq analysis precisely mapped many transcript boundaries and identified 75 new genes.
Findings: We now report a much deeper analysis using the SOLiD™ technology combining RNA-Seq of the Mimivirus transcriptome during the infectious cycle (202.4 Million reads), and a complete genome re-sequencing (45.3 Million reads). This study corrected the genome sequence and identified several single nucleotide polymorphisms. Our results also provided clear evidence of previously overlooked transcription units, including an important RNA polymerase subunit distantly related to Euryarchea homologues. The total Mimivirus gene count is now 1018, 11% greater than the original annotation.
Conclusions: This study highlights the huge progress brought about by ultra-deep sequencing for the comprehensive annotation of virus genomes, opening the door to a complete one-nucleotide resolution level description of their transcriptional activity, and to the realistic modeling of the viral genome expression at the ultimate molecular level. This work also illustrates the need to go beyond bioinformatics-only approaches for the annotation of short protein and non-coding genes in viral genomes.
Figures
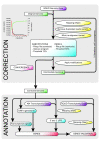
Figure 1
Flow chart of the Mimivirus genome correction pipeline. The upper panel illustrates the correction procedure and the lower panel the annotation method. Colors are used for clarity: datasets are in purple, genomes are in green, sequence manipulations (mapping, duplicate removal, or modifications) are in yellow, computation steps are in blue and genes in red. The upper left graph represents the decrease in substitutions (in red) and indels (in black) identified during the iterative genome correction process, together with the increase in the total number of reads (in green) mapped to genome.
Figure 2
Discovery of a component of the Mimivirus transcription apparatus. A) Mimivirus genome browser (URL:
http://www.igs.cnrs-mrs.fr/mimivirus/
) screenshot showing the newly discovered component of the transcription apparatus (R357b) in its genomic context. Three informative tracks are displayed: the protein coding genes, the late gene expression signals, and the gene expression data from the SOLiD™ RNA-seq experiment. Transcriptome data is shown at each genomic position (for each of the 9 samples) going from white (not expressed) to red (highly expressed) in the forward strand, and white to blue (highly expressed) in the reverse strand. B) Protein sequence alignment of the Mimivirus R357b gene and the most similar homologous sequences from the giant virus CroV and the two archea Methanocella paludicola and Ferroplasma acidarmanus.
Similar articles
- mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus.
Legendre M, Audic S, Poirot O, Hingamp P, Seltzer V, Byrne D, Lartigue A, Lescot M, Bernadac A, Poulain J, Abergel C, Claverie JM. Legendre M, et al. Genome Res. 2010 May;20(5):664-74. doi: 10.1101/gr.102582.109. Epub 2010 Apr 1. Genome Res. 2010. PMID: 20360389 Free PMC article. - Mimivirus and its virophage.
Claverie JM, Abergel C. Claverie JM, et al. Annu Rev Genet. 2009;43:49-66. doi: 10.1146/annurev-genet-102108-134255. Annu Rev Genet. 2009. PMID: 19653859 Review. - Viruses with more than 1,000 genes: Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, and reannotation of Mimivirus genes.
Colson P, Yutin N, Shabalina SA, Robert C, Fournous G, La Scola B, Raoult D, Koonin EV. Colson P, et al. Genome Biol Evol. 2011;3:737-42. doi: 10.1093/gbe/evr048. Epub 2011 Jun 24. Genome Biol Evol. 2011. PMID: 21705471 Free PMC article. - Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae.
Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. Arslan D, et al. Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17486-91. doi: 10.1073/pnas.1110889108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987820 Free PMC article. - Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.
Iyer LM, Balaji S, Koonin EV, Aravind L. Iyer LM, et al. Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review.
Cited by
- Mimivirus encodes a multifunctional primase with DNA/RNA polymerase, terminal transferase and translesion synthesis activities.
Gupta A, Lad SB, Ghodke PP, Pradeepkumar PI, Kondabagil K. Gupta A, et al. Nucleic Acids Res. 2019 Jul 26;47(13):6932-6945. doi: 10.1093/nar/gkz236. Nucleic Acids Res. 2019. PMID: 31001622 Free PMC article. - Exploration of the propagation of transpovirons within Mimiviridae reveals a unique example of commensalism in the viral world.
Jeudy S, Bertaux L, Alempic JM, Lartigue A, Legendre M, Belmudes L, Santini S, Philippe N, Beucher L, Biondi EG, Juul S, Turner DJ, Couté Y, Claverie JM, Abergel C. Jeudy S, et al. ISME J. 2020 Mar;14(3):727-739. doi: 10.1038/s41396-019-0565-y. Epub 2019 Dec 10. ISME J. 2020. PMID: 31822788 Free PMC article. - Taxon Richness of "Megaviridae" Exceeds those of Bacteria and Archaea in the Ocean.
Mihara T, Koyano H, Hingamp P, Grimsley N, Goto S, Ogata H. Mihara T, et al. Microbes Environ. 2018 Jul 4;33(2):162-171. doi: 10.1264/jsme2.ME17203. Epub 2018 May 25. Microbes Environ. 2018. PMID: 29806626 Free PMC article. - A novel and diverse family of filamentous DNA viruses associated with parasitic wasps.
Guinet B, Leobold M, Herniou EA, Bloin P, Burlet N, Bredlau J, Navratil V, Ravallec M, Uzbekov R, Kester K, Gundersen Rindal D, Drezen JM, Varaldi J, Bézier A. Guinet B, et al. Virus Evol. 2024 Mar 2;10(1):veae022. doi: 10.1093/ve/veae022. eCollection 2024. Virus Evol. 2024. PMID: 38617843 Free PMC article. - Comparative genomic and biochemical analyses identify a collagen galactosylhydroxylysyl glucosyltransferase from Acanthamoeba polyphaga mimivirus.
Wu W, Kim JS, Bailey AO, Russell WK, Richards SJ, Chen T, Chen T, Chen Z, Liang B, Yamauchi M, Guo H. Wu W, et al. Sci Rep. 2022 Oct 7;12(1):16806. doi: 10.1038/s41598-022-21197-1. Sci Rep. 2022. PMID: 36207453 Free PMC article.
References
- Legendre M, Audic S, Poirot O, Hingamp P, Seltzer V, Byrne D, Lartigue A, Lescot M, Bernadac A, Poulain J, Abergel C, Claverie JM. mRNA deep sequencing reveals 75 new genes and a complex transcriptional landscape in Mimivirus. Genome Res. 2010;20:664–674. doi: 10.1101/gr.102582.109. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous