The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes - PubMed (original) (raw)
The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes
Mark W J van Passel et al. PLoS One. 2011.
Abstract
Background: The human gastrointestinal tract contains a complex community of microbes, fulfilling important health-promoting functions. However, this vast complexity of species hampers the assignment of responsible organisms to these functions. Recently, Akkermansia muciniphila, a new species from the deeply branched phylum Verrucomicrobia, was isolated from the human intestinal tract based on its capacity to efficiently use mucus as a carbon and nitrogen source. This anaerobic resident is associated with the protective mucus lining of the intestines.
Methodology/principal findings: In order to uncover the functional potential of A. muciniphila, its genome was sequenced and annotated. It was found to contain numerous candidate mucinase-encoding genes, but lacking genes encoding canonical mucus-binding domains. Numerous phage-associated sequences found throughout the genome indicate that viruses have played an important part in the evolution of this species. Furthermore, we mined 37 GI tract metagenomes for the presence, and genetic diversity of Akkermansia sequences. Out of 37, eleven contained 16S ribosomal RNA gene sequences that are >95% identical to that of A. muciniphila. In addition, these libraries were found to contain large amounts of Akkermansia DNA based on average nucleotide identity scores, which indicated in one subject co-colonization by different Akkermansia phylotypes. An additional 12 libraries also contained Akkermansia sequences, making a total of ∼16 Mbp of new Akkermansia pangenomic DNA. The relative abundance of Akkermansia DNA varied between <0.01% to nearly 4% of the assembled metagenomic reads. Finally, by testing a large collection of full length 16S sequences, we find at least eight different representative species in the genus Akkermansia.
Conclusions/significance: These large repositories allow us to further mine for genetic heterogeneity and species diversity in the genus Akkermansia, providing novel insight towards the functionality of this abundant inhabitant of the human intestinal tract.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
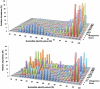
Figure 1. Average nucleotide identity (ANI) distributions for the 11 metagenomic libraries in which we found _Akkermansia_-like 16S rRNA sequences (1A) and for the 12 metagenomic libraries in which we did not find _Akkermansia_-like 16S rRNA sequences (1B), but which did show numerous hits with the A. muciniphila genome with nucleotide identity scores above 90%.
Please note the distinctive distribution of ANI values of libraries B, A and MH6. Relative abundance scores (z-axis) are calculated for the total number of contigs per metagenome that show nucleotide identity scores >75%.
Similar articles
- Genomic convergence between Akkermansia muciniphila in different mammalian hosts.
Geerlings SY, Ouwerkerk JP, Koehorst JJ, Ritari J, Aalvink S, Stecher B, Schaap PJ, Paulin L, de Vos WM, Belzer C. Geerlings SY, et al. BMC Microbiol. 2021 Oct 29;21(1):298. doi: 10.1186/s12866-021-02360-6. BMC Microbiol. 2021. PMID: 34715771 Free PMC article. - Genome-Scale Model and Omics Analysis of Metabolic Capacities of Akkermansia muciniphila Reveal a Preferential Mucin-Degrading Lifestyle.
Ottman N, Davids M, Suarez-Diez M, Boeren S, Schaap PJ, Martins Dos Santos VAP, Smidt H, Belzer C, de Vos WM. Ottman N, et al. Appl Environ Microbiol. 2017 Aug 31;83(18):e01014-17. doi: 10.1128/AEM.01014-17. Print 2017 Sep 15. Appl Environ Microbiol. 2017. PMID: 28687644 Free PMC article. - Genomic diversity and ecology of human-associated Akkermansia species in the gut microbiome revealed by extensive metagenomic assembly.
Karcher N, Nigro E, Punčochář M, Blanco-Míguez A, Ciciani M, Manghi P, Zolfo M, Cumbo F, Manara S, Golzato D, Cereseto A, Arumugam M, Bui TPN, Tytgat HLP, Valles-Colomer M, de Vos WM, Segata N. Karcher N, et al. Genome Biol. 2021 Jul 14;22(1):209. doi: 10.1186/s13059-021-02427-7. Genome Biol. 2021. PMID: 34261503 Free PMC article. - A next generation probiotic, Akkermansia muciniphila.
Zhai Q, Feng S, Arjan N, Chen W. Zhai Q, et al. Crit Rev Food Sci Nutr. 2019;59(19):3227-3236. doi: 10.1080/10408398.2018.1517725. Epub 2018 Oct 29. Crit Rev Food Sci Nutr. 2019. PMID: 30373382 Review. - Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How?
Geerlings SY, Kostopoulos I, de Vos WM, Belzer C. Geerlings SY, et al. Microorganisms. 2018 Jul 23;6(3):75. doi: 10.3390/microorganisms6030075. Microorganisms. 2018. PMID: 30041463 Free PMC article. Review.
Cited by
- The role of the gut microbiota in nutrition and health.
Flint HJ, Scott KP, Louis P, Duncan SH. Flint HJ, et al. Nat Rev Gastroenterol Hepatol. 2012 Sep 4;9(10):577-89. doi: 10.1038/nrgastro.2012.156. eCollection 2012 Oct. Nat Rev Gastroenterol Hepatol. 2012. PMID: 22945443 Review. - Alterations in the Abundance and Co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the Colonic Mucosa of Inflammatory Bowel Disease Subjects.
Lopez-Siles M, Enrich-Capó N, Aldeguer X, Sabat-Mir M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Lopez-Siles M, et al. Front Cell Infect Microbiol. 2018 Sep 7;8:281. doi: 10.3389/fcimb.2018.00281. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30245977 Free PMC article. - Akkermansia muciniphila Ameliorates Clostridioides difficile Infection in Mice by Modulating the Intestinal Microbiome and Metabolites.
Wu Z, Xu Q, Gu S, Chen Y, Lv L, Zheng B, Wang Q, Wang K, Wang S, Xia J, Yang L, Bian X, Jiang X, Zheng L, Li L. Wu Z, et al. Front Microbiol. 2022 May 18;13:841920. doi: 10.3389/fmicb.2022.841920. eCollection 2022. Front Microbiol. 2022. PMID: 35663882 Free PMC article. - Utilization Efficiency of Human Milk Oligosaccharides by Human-Associated Akkermansia Is Strain Dependent.
Luna E, Parkar SG, Kirmiz N, Hartel S, Hearn E, Hossine M, Kurdian A, Mendoza C, Orr K, Padilla L, Ramirez K, Salcedo P, Serrano E, Choudhury B, Paulchakrabarti M, Parker CT, Huynh S, Cooper K, Flores GE. Luna E, et al. Appl Environ Microbiol. 2022 Jan 11;88(1):e0148721. doi: 10.1128/AEM.01487-21. Epub 2021 Oct 20. Appl Environ Microbiol. 2022. PMID: 34669436 Free PMC article. - Akkermansia and Microbial Degradation of Mucus in Cats and Dogs: Implications to the Growing Worldwide Epidemic of Pet Obesity.
Garcia-Mazcorro JF, Minamoto Y, Kawas JR, Suchodolski JS, de Vos WM. Garcia-Mazcorro JF, et al. Vet Sci. 2020 Apr 15;7(2):44. doi: 10.3390/vetsci7020044. Vet Sci. 2020. PMID: 32326394 Free PMC article. Review.
References
- Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous