Genetic dissection of the Gpnmb network in the eye - PubMed (original) (raw)
. 2011 Jun 13;52(7):4132-42.
doi: 10.1167/iovs.10-6493.
Xusheng Wang, Matthew Pullen, Huaijin Guan, Hui Chen, Shwetapadma Sahu, Bing Zhang, Hao Chen, Robert W Williams, Eldon E Geisert, Lu Lu, Monica M Jablonski
Affiliations
- PMID: 21398278
- PMCID: PMC3175941
- DOI: 10.1167/iovs.10-6493
Genetic dissection of the Gpnmb network in the eye
Hong Lu et al. Invest Ophthalmol Vis Sci. 2011.
Abstract
Purpose: To use a systematic genetics approach to investigate the regulation of Gpnmb, a gene that contributes to pigmentary dispersion syndrome (PDS) and pigmentary glaucoma (PG) in the DBA/2J (D2) mouse.
Methods: Global patterns of gene expression were studied in whole eyes of a large family of BXD mouse strains (n = 67) generated by crossing the PDS- and PG-prone parent (DBA/2J) with a resistant strain (C57BL/6J). Quantitative trait locus (eQTL) mapping methods and gene set analysis were used to evaluate Gpnmb coexpression networks in wild-type and mutant cohorts.
Results: The level of Gpnmb expression was associated with a highly significant cis-eQTL at the location of the gene itself. This autocontrol of Gpnmb is likely to be a direct consequence of the known premature stop codon in exon 4. Both gene ontology and coexpression network analyses demonstrated that the mutation in Gpnmb radically modified the set of genes with which Gpnmb expression is correlated. The covariates of wild-type Gpnmb are involved in biological processes including melanin synthesis and cell migration, whereas the covariates of mutant Gpnmb are involved in the biological processes of posttranslational modification, stress activation, and sensory processing.
Conclusions: These results demonstrated that a systematic genetics approach provides a powerful tool for constructing coexpression networks that define the biological process categories within which similarly regulated genes function. The authors showed that the R150X mutation in Gpnmb dramatically modified its list of genetic covariates, which may explain the associated ocular pathology.
Figures
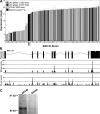
Figure 1.
Expression levels of Gpnmb across strains. (A) Rank-ordered mean Gpnmb levels across 67 BXD RI strains, their parental strains, and F1 crosses. N, two male and two female mice per strain. Values denote normalized relative expression levels on a log2 scale (mean ± SEM). All but two RI strains (BXD40 and BXD42) generated by the Jackson Laboratory (JAX) have high levels of Gpnmb. The RI lines we generated at UTHSC express both high and low levels of Gpnmb. (B) RNA-seq data showing the level of Gpnmb across all 11 exons from D2 (bottom, white bar) and B6 (middle, black bar) parental strains. The level of Gpnmb message was greater in the B6 mouse across the entire transcript. (C) Whole eye lysates from BXD lines with high and low mRNA expression levels underwent Western blot analysis with an anti-GPNMB antibody. This representative blot shows the absence GPNMB protein at the expected molecular weight in lines with low mRNA expression levels.
Figure 2.
Graphic illustration of LRS scores for Gpnmb expression in eyes of BXD mice. A significant eQTL for Gpnmb is present on chromosome 6 at the location of the gene itself. Dark gray trace: LRS scores across the genome; horizontal lines: transcript-specific significance thresholds for significant (LRS∼17) and suggestive (LRS∼10) based on results of 1000 permutations of the original trait data.
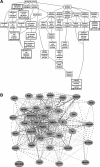
Figure 3.
Genetic associations with wild-type Gpnmb. (A) In BXD RI mice with wild-type Gpnmb, GO enrichment analysis illustrates that most biological processes to which transcripts correlated with Gpnmb expression belong include cell motion, cell migration, and melanin synthesis. Enriched categories reaching statistical significance are indicated with a bold outline. (B) Genetic coexpression network generated from genes correlated with wild-type Gpnmb shows that the expression level of Gpnmb was directly linked to five nodes. Other correlations with Gpnmb within the network occurred through intermediary transcripts. Each transcript is shown as a node, and the Pearson correlation coefficient between nodes is indicated by a line. Bold lines: coefficients of 1.0 to 0.7; normal lines: coefficients of 0.7 to 0.5; dashed lines: coefficients of 0.5 to 0.0.
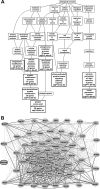
Figure 4.
Genetic associations with mutant Gpnmb. (A) In BXD RI mice with mutant Gpnmb, GO enrichment analysis illustrates that the majority of biological processes to which transcripts correlated with Gpnmb expression belong include posttranslational protein modifications, stress-activated kinase signaling, and visual perception. Enriched categories reaching statistical significance are indicated with a bold outline. (B) A genetic coexpression network generated from genes correlated with mutant Gpnmb shows that Gpnmb was directly linked to 10 nodes in the network. All other interactions occurred through intermediaries. Each transcript is shown as a node, and the Pearson correlation coefficient between nodes is indicated by a line. Bold lines: coefficients of 1.0 to 0.7; normal lines: coefficients of 0.7 to 0.5; dashed lines: coefficients of 0.5 to 0.0.
Figure 5.
Heat maps of Gpnmb covariates. (A) In BXD RI mice with wild-type Gpnmb, tight bands of correlates are present on chromosomes 5, 9, and 19, and more dispersed bands are present on chromosomes 8 and 10. (B) In RI strains with mutant Gpnmb, the heat map shows tight bands of correlates on chromosomes 4, 5, and 8.
Similar articles
- Investigating a downstream gene of Gpnmb using the systems genetics method.
Lu Y, Zhou D, Lu H, Xu F, Yue J, Tong J, Lu L. Lu Y, et al. Mol Vis. 2019 Apr 23;25:222-236. eCollection 2019. Mol Vis. 2019. PMID: 31057322 Free PMC article. - Iris transillumination defect and its gene modulators do not correlate with intraocular pressure in the BXD family of mice.
Lu H, Lu L, Williams RW, Jablonski MM. Lu H, et al. Mol Vis. 2016 Mar 4;22:224-33. eCollection 2016. Mol Vis. 2016. PMID: 27011731 Free PMC article. - Complex interactions of Tyrp1 in the eye.
Lu H, Li L, Watson ER, Williams RW, Geisert EE, Jablonski MM, Lu L. Lu H, et al. Mol Vis. 2011;17:2455-68. Epub 2011 Sep 22. Mol Vis. 2011. PMID: 21976956 Free PMC article. - The genetics of pigment dispersion syndrome and pigmentary glaucoma.
Lascaratos G, Shah A, Garway-Heath DF. Lascaratos G, et al. Surv Ophthalmol. 2013 Mar-Apr;58(2):164-75. doi: 10.1016/j.survophthal.2012.08.002. Epub 2012 Dec 6. Surv Ophthalmol. 2013. PMID: 23218808 Review. - [Glaucoma genetics].
Romanenko IA. Romanenko IA. Voen Med Zh. 2009 Jun;330(6):46-50. Voen Med Zh. 2009. PMID: 19791688 Review. Russian. No abstract available.
Cited by
- Analyses of differentially expressed genes after exposure to acute stress, acute ethanol, or a combination of both in mice.
Baker JA, Li J, Zhou D, Yang M, Cook MN, Jones BC, Mulligan MK, Hamre KM, Lu L. Baker JA, et al. Alcohol. 2017 Feb;58:139-151. doi: 10.1016/j.alcohol.2016.08.008. Epub 2016 Dec 16. Alcohol. 2017. PMID: 28027852 Free PMC article. - Systems Genetics of Optic Nerve Axon Necrosis During Glaucoma.
Stiemke AB, Sah E, Simpson RN, Lu L, Williams RW, Jablonski MM. Stiemke AB, et al. Front Genet. 2020 Feb 27;11:31. doi: 10.3389/fgene.2020.00031. eCollection 2020. Front Genet. 2020. PMID: 32174956 Free PMC article. - Genetic modulation of the iris transillumination defect: a systems genetics analysis using the expanded family of BXD glaucoma strains.
Swaminathan S, Lu H, Williams RW, Lu L, Jablonski MM. Swaminathan S, et al. Pigment Cell Melanoma Res. 2013 Jul;26(4):487-98. doi: 10.1111/pcmr.12106. Epub 2013 May 13. Pigment Cell Melanoma Res. 2013. PMID: 23582180 Free PMC article. - Exploring Early-Stage Retinal Neurodegeneration in Murine Pigmentary Glaucoma: Insights From Gene Networks and miRNA Regulation Analyses.
Gu Q, Kumar A, Hook M, Xu F, Bajpai AK, Starlard-Davenport A, Yue J, Jablonski MM, Lu L. Gu Q, et al. Invest Ophthalmol Vis Sci. 2023 Sep 1;64(12):25. doi: 10.1167/iovs.64.12.25. Invest Ophthalmol Vis Sci. 2023. PMID: 37707836 Free PMC article. - A Spontaneous Mutation in Taar1 Impacts Methamphetamine-Related Traits Exclusively in DBA/2 Mice from a Single Vendor.
Reed C, Baba H, Zhu Z, Erk J, Mootz JR, Varra NM, Williams RW, Phillips TJ. Reed C, et al. Front Pharmacol. 2018 Jan 22;8:993. doi: 10.3389/fphar.2017.00993. eCollection 2017. Front Pharmacol. 2018. PMID: 29403379 Free PMC article.
References
- Ritch R, Shields MB, Krupin T. The Glaucomas, Clinical Science. 2nd ed St Louis, MO: Mosby-Year Book; 1996
- Leske MC. The epidemiology of open-angle glaucoma: a review. Am J Epidemiol. 1983;118:166–191 - PubMed
- Wiggs JL. Genetic etiologies of glaucoma. Arch Ophthalmol. 2007;125:30–37 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AA014425/AA/NIAAA NIH HHS/United States
- AA017590/AA/NIAAA NIH HHS/United States
- P20 DA021131/DA/NIDA NIH HHS/United States
- U01 AA017590/AA/NIAAA NIH HHS/United States
- R01 EY017841/EY/NEI NIH HHS/United States
- EY021200/EY/NEI NIH HHS/United States
- EY017841/EY/NEI NIH HHS/United States
- P30 EY013080/EY/NEI NIH HHS/United States
- P30EY013080/EY/NEI NIH HHS/United States
- R01 EY021200/EY/NEI NIH HHS/United States
- DA021131/DA/NIDA NIH HHS/United States
- U01 AA014425/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous