Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells - PubMed (original) (raw)
. 2011 May 19;473(7347):389-93.
doi: 10.1038/nature09934. Epub 2011 Mar 30.
Affiliations
- PMID: 21451524
- PMCID: PMC3539771
- DOI: 10.1038/nature09934
Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells
Hao Wu et al. Nature. 2011.
Abstract
Epigenetic modification of the mammalian genome by DNA methylation (5-methylcytosine) has a profound impact on chromatin structure, gene expression and maintenance of cellular identity. The recent demonstration that members of the Ten-eleven translocation (Tet) family of proteins can convert 5-methylcytosine to 5-hydroxymethylcytosine raised the possibility that Tet proteins are capable of establishing a distinct epigenetic state. We have recently demonstrated that Tet1 is specifically expressed in murine embryonic stem (ES) cells and is required for ES cell maintenance. Using chromatin immunoprecipitation coupled with high-throughput DNA sequencing, here we show in mouse ES cells that Tet1 is preferentially bound to CpG-rich sequences at promoters of both transcriptionally active and Polycomb-repressed genes. Despite an increase in levels of DNA methylation at many Tet1-binding sites, Tet1 depletion does not lead to downregulation of all the Tet1 targets. Interestingly, although Tet1-mediated promoter hypomethylation is required for maintaining the expression of a group of transcriptionally active genes, it is also involved in repression of Polycomb-targeted developmental regulators. Tet1 contributes to silencing of this group of genes by facilitating recruitment of PRC2 to CpG-rich gene promoters. Thus, our study not only establishes a role for Tet1 in modulating DNA methylation levels at CpG-rich promoters, but also reveals a dual function of Tet1 in promoting transcription of pluripotency factors as well as participating in the repression of Polycomb-targeted developmental regulators.
Figures
Figure 1. Tet1 is enriched at genomic regions with high-density CpG dinucleotides
a, Genome-wide occupancy of Tet1 at all annotated gene promoters in ES cells (black, CpG-rich genes; red, CpG-poor genes). The enrichment of Tet1 binding was determined by ChIP-seq analysis. Average Tet1 binding measured by −log10 (peak P values) in 200-bp bins is shown within genomic regions covering 5 kb up- and downstream of TSSs. b, Enrichment of Tet1 (purple), Kdm2a (orange) and H3K4me3 (green) measured by ChIP-seq at representative genes in ES cells (black, CpG-rich; red, CpG-poor). ChIP-seq data are shown in reads per million with the _y_-axis floor set to 0.5 reads per million. Genomic regions with statistically significant enrichment of Tet1 binding (measured by log10 (peak P values); P < 10−8) are also indicated. c, Heatmap representation of genomic regions with high-density CpG sites (CpG islands), binding profiles of Tet1, Kdm2a and H3K4me3 in ES cells at all annotated mouse genes promoters (5 kb flanking TSSs of Refseq genes). The heatmap is rank-ordered from genes with CpG islands of longest length to no CpG islands within 5-kb genomic regions flanking TSSs. The presence of CpG islands is shown in colour (blue, present; white, absent). ChIP-seq enrichment was measured by −log10 (peak P values) and is shown by colour scale. The following colour scales (white, no enrichment; blue, high enrichment) were used for Tet1/Kdm2a and H3K4me3 respectively: (0, 50) and (0, 200). d, A DNA motif that is enriched in Tet1-bound loci in ES cells.
Figure 2. Tet1 maintains a DNA hypomethylated state at Tet1-bound regions
a, The distribution frequency of regions enriched with DNA methylation is shown for Tet1-bound (blue) and unbound (red) gene promoters (left) or CpG islands (right) in mouse ES cells. b, Heatmap representation of CpG islands and the changes in DNA methylation (5mC) in response to Tet1 depletion. The DNA methylation gained after Tet1 depletion was calculated by deduction of 5mC levels in control knockdown (Con KD) from that in Tet1 knockdown (Tet1 KD). c, Changes in 5mC levels in response to Tet1 knockdown are shown for both CpG-rich and CpG-poor gene promoters. Note that proximal promoters and 5′ intragenic regions of CpG-rich genes are associated with a higher increase in 5mC levels as compared to those of CpG-poor genes in response to Tet1 depletion. d, An increase in 5mC levels in response to Tet1 depletion is specifically enriched at the centre of Tet1-binding loci. Changes in 5mC levels between control knockdown and Tet1 knockdown ES cells were determined by co-hybridizing and analysing genomic DNA from control knockdown and Tet1 knockdown cells on the whole-genome tiling microarrays.
Figure 3. Tet1 binds to and functions in both repressed (bivalent) and actively transcribed (H3K4me3-only) genes
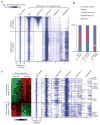
a, Heatmap representation of genomic regions with high-density CpG sites (CpG islands), binding profile of Tet1, and major histone modifications (H3K4me1 (ref. 12), H3K4me3, H3K27me3 and H3K36me3 (ref. 13)) in mouse ES cells at indicated Tet1 target genes (5 kb flanking TSSs). The heatmap is rank-ordered from genes with highest H3K27me3 enrichment to no H3K27me3 within 5-kb genomic regions flanking TSSs. The following colour scales (white, no enrichment; blue, high enrichment) were used for Tet1/H3K27me3/H3K4me1/ H3K36me3 and H3K4me3 respectively: (0, 50) and (0, 200). WT, wild type. b, Relative percentage of genes with different chromatin states shown for all genes, Tet1-bound and unbound genes. c, Heatmap representation of differentially expressed Tet1 targets between control knockdown and Tet1 knockdown mouse ES cells. Note that Tet1-repressed targets are preferentially associated with bivalent chromatin states, whereas Tet1-activated targets are generally H3K4me3-only genes.
Figure 4. Tet1 is required for chromatin binding of PRC2 in mouse ES cells
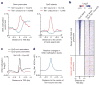
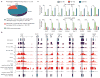
a, Tet1 depletion affects the binding of PRC2 to the majority of its targets. PRC2-binding sites are divided into three groups (Tet1/PRC2 co-bound Tet1 dependent, Tet1 independent and PRC2-only bound). b, Shown are Tet1, Ezh2 and Suz12 (ref. 21), and H3K27me3 (ref. 13) occupancy, and the effect of Tet1 depletion on Ezh2 occupancy and 5mC levels at seven representative Tet1-repressed bivalent targets. Regions associated with significant changes in Ezh2 occupancy between control and _Tet1_-depleted ES cells were measured by whole-genome tiling microarrays. Genomic regions that are further examined by locus-specific ChIP–qPCR in c are shaded. c, ChIP–qPCR analysis of Tet1 (top panels) and Ezh2 (bottom panels) occupancy at the promoters of eight representative Tet1-repressed targets in control (Con KD), _Tet1_-depleted (Tet1 KD) and _Ezh2_-depleted (Ezh2 KD) ES cells. Error bars represents standard deviation determined from duplicate experiments.
Comment in
- Epigenetics: Tet proteins in the limelight.
Véron N, Peters AH. Véron N, et al. Nature. 2011 May 19;473(7347):293-4. doi: 10.1038/473293a. Nature. 2011. PMID: 21593859 No abstract available.
Similar articles
- TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity.
Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. Williams K, et al. Nature. 2011 May 19;473(7347):343-8. doi: 10.1038/nature10066. Epub 2011 Apr 13. Nature. 2011. PMID: 21490601 Free PMC article. - Epigenetics: Tet proteins in the limelight.
Véron N, Peters AH. Véron N, et al. Nature. 2011 May 19;473(7347):293-4. doi: 10.1038/473293a. Nature. 2011. PMID: 21593859 No abstract available. - Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells.
Neri F, Incarnato D, Krepelova A, Rapelli S, Pagnani A, Zecchina R, Parlato C, Oliviero S. Neri F, et al. Genome Biol. 2013 Aug 29;14(8):R91. doi: 10.1186/gb-2013-14-8-r91. Genome Biol. 2013. PMID: 23987249 Free PMC article. - DNA methylation: TET proteins-guardians of CpG islands?
Williams K, Christensen J, Helin K. Williams K, et al. EMBO Rep. 2011 Dec 23;13(1):28-35. doi: 10.1038/embor.2011.233. EMBO Rep. 2011. PMID: 22157888 Free PMC article. Review. - Uncovering the role of 5-hydroxymethylcytosine in the epigenome.
Branco MR, Ficz G, Reik W. Branco MR, et al. Nat Rev Genet. 2011 Nov 15;13(1):7-13. doi: 10.1038/nrg3080. Nat Rev Genet. 2011. PMID: 22083101 Review.
Cited by
- 5-Hydroxymethylcytosine: Far Beyond the Intermediate of DNA Demethylation.
Zheng K, Lyu Z, Chen J, Chen G. Zheng K, et al. Int J Mol Sci. 2024 Nov 2;25(21):11780. doi: 10.3390/ijms252111780. Int J Mol Sci. 2024. PMID: 39519332 Free PMC article. Review. - Hypermethylation of CDKN2A CpG island drives resistance to PRC2 inhibitors in SWI/SNF loss-of-function tumors.
Wang X, Wang Y, Xie M, Ma S, Zhang Y, Wang L, Ge Y, Li G, Zhao M, Chen S, Yan C, Zhang H, Sun W. Wang X, et al. Cell Death Dis. 2024 Nov 5;15(11):794. doi: 10.1038/s41419-024-07109-3. Cell Death Dis. 2024. PMID: 39500892 Free PMC article. - The Role of DNMT Methyltransferases and TET Dioxygenases in the Maintenance of the DNA Methylation Level.
Davletgildeeva AT, Kuznetsov NA. Davletgildeeva AT, et al. Biomolecules. 2024 Sep 4;14(9):1117. doi: 10.3390/biom14091117. Biomolecules. 2024. PMID: 39334883 Free PMC article. Review. - TET enzyme driven epigenetic reprogramming in early embryos and its implication on long-term health.
Montgomery T, Uh K, Lee K. Montgomery T, et al. Front Cell Dev Biol. 2024 Aug 1;12:1358649. doi: 10.3389/fcell.2024.1358649. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39149518 Free PMC article. Review. - Loss of Tet hydroxymethylase activity causes mouse embryonic stem cell differentiation bias and developmental defects.
Wang M, Wang L, Huang Y, Qiao Z, Yi S, Zhang W, Wang J, Yang G, Cui X, Kou X, Zhao Y, Wang H, Jiang C, Gao S, Chen J. Wang M, et al. Sci China Life Sci. 2024 Oct;67(10):2132-2148. doi: 10.1007/s11427-024-2631-x. Epub 2024 Jul 5. Sci China Life Sci. 2024. PMID: 39037697
References
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nature Rev Genet. 2008;2008:129–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- ImNIH/Intramural NIH HHS/United States
- R01 GM068804/GM/NIGMS NIH HHS/United States
- GM68804/GM/NIGMS NIH HHS/United States
- R56 MH082068/MH/NIMH NIH HHS/United States
- R56MH082068/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases