Protein inhibitor of activated STAT3 expression in lung cancer - PubMed (original) (raw)
Protein inhibitor of activated STAT3 expression in lung cancer
Amy Kluge et al. Mol Oncol. 2011 Jun.
Abstract
Protein Inhibitor of Activated Signal Transducer and Activators of Transcription 3 (PIAS3) is an endogenous inhibitor of STAT3 transcriptional activity. We have previously demonstrated the concentration-dependent negative regulatory effect of PIAS3 on STAT3 signaling and its capacity to decrease lung cancer proliferation and synergize with epidermal growth factor inhibition. We now investigate PIAS3 expression in both non-small cell lung cancer (NSCLC) cell lines and human resected NSCLC specimens. We also investigated the mechanism by which some lung cancers have significantly decreased PIAS3 expression. Expression of PIAS3 is variable in lung cancer cells lines with 2 of 3 squamous cell carcinoma (SCC) cell lines having no or little PIAS3 protein expression. Similarly, the majority of human SCCs of the lung lack PIAS3 expression by immunohistochemistry; this despite the finding that SCCs have significantly higher levels of PIAS3 mRNA compared to adenocarcinomas. High PIAS3 expression generally correlates with decreased phosphorylated STAT3 in both SCC cell lines and human specimens compatible with the negative regulatory effect of this protein on STAT3 signaling. To investigate this variable expression of PIAS3 we first performed sequencing of the PIAS3 gene that demonstrated single nucleotide polymorphisms but no mutations. Exposure of lung cancer cells to 5-azacytidine and trichostatin A results in a significant increase in PIAS3 mRNA and protein expression. However, methylation-specific PCR demonstrates a lack of CpG island methylation in the promoter region of PIAS3. Exposure of cells to an agent blocking proteosomal degradation results in a significant increase in PIAS3. Our data thus shows that SCC of the lung commonly lacks PIAS3 protein expression and that post-translational modifications may explain this finding in some cases. PIAS3 is a potential therapeutic molecule to target STAT3 pathway in lung cancer.
Copyright © 2011 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
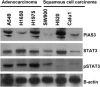
Figure 1
Western blot demonstrates variable expression of PIAS3 in NSCLC cell lines. Phosphorylated STAT3 expression and STAT3 expression were also evaluated. β‐actin was used as a protein loading control.
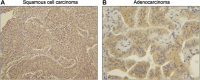
Figure 2
Immunohistochemistry for PIAS3 in resected lung cancer. Panel A demonstrates a squamous cell carcinoma with no PIAS3 staining and panel B demonstrates an adenocarcinoma with 3+ staining.
Figure 3
Single nucleotide polymorphisms found in A549 sense sequence, change from C/T or G/T. Verified the SNP changes by sequencing the A549 antisense and confirmed the G/A and C/A changes. These C/T and G/T SNPs were found in other sequenced cell lines, both adenocarcinomas and squamous cell carcinomas, as well as in NuLi cells, an immortalized normal lung epithelial cell line.
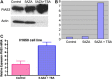
Figure 4
A‐ PIAS3 expression and β‐actin expression in H1650 cells. Cells were treated with 5AZA for 48 h followed by a 24 h treatment with TSA. B‐ Relative expression of PIAS3 standardized to β‐actin expression shows that 5AZA increased PIAS3 protein but that the combination of 5AZA and TSA increased expression about 8‐fold higher. C‐ A ∼3‐fold increase in relative PIAS3 mRNA expression in H1650 cells can be seen after exposure to 5AZA + TSA by real‐time quantitative PCR.
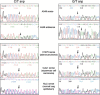
Figure 5
Localization of PIAS3 qMSP assays on Chromosome 1: Transcription Start Site (TSS) is shown as pink vertical lines, the green box represents the first exon, and the open box represents the PIAS3 promoter region. Open red boxes represent location of PIAS3 qMSP assays used for methylation profiling.
Figure 6
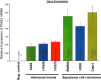
Relative mRNA expression of adenocarcinoma v. squamous cell carcinoma cell lines.
Figure 7
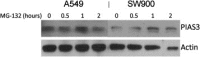
Post‐translational proteosomal degradation by MG‐132 in adenocarcinoma v. squamous cell carcinoma cell lines.
Similar articles
- PIAS3 expression in squamous cell lung cancer is low and predicts overall survival.
Abbas R, McColl KS, Kresak A, Yang M, Chen Y, Fu P, Wildey G, Dowlati A. Abbas R, et al. Cancer Med. 2015 Mar;4(3):325-32. doi: 10.1002/cam4.372. Epub 2015 Jan 9. Cancer Med. 2015. PMID: 25573684 Free PMC article. - Cooperative interaction between protein inhibitor of activated signal transducer and activator of transcription-3 with epidermal growth factor receptor blockade in lung cancer.
Kluge A, Dabir S, Kern J, Nethery D, Halmos B, Ma P, Dowlati A. Kluge A, et al. Int J Cancer. 2009 Oct 1;125(7):1728-34. doi: 10.1002/ijc.24553. Int J Cancer. 2009. PMID: 19569236 Free PMC article. - Expression of STAT5, COX-2 and PIAS3 in correlation with NSCLC histhopathological features.
Pastuszak-Lewandoska D, Domańska D, Czarnecka KH, Kordiak J, Migdalska-Sęk M, Nawrot E, Kiszałkiewicz J, Antczak A, Górski P, Brzeziańska E. Pastuszak-Lewandoska D, et al. PLoS One. 2014 Aug 19;9(8):e104265. doi: 10.1371/journal.pone.0104265. eCollection 2014. PLoS One. 2014. PMID: 25137041 Free PMC article. - The Roles of Post-Translational Modifications in STAT3 Biological Activities and Functions.
Tesoriere A, Dinarello A, Argenton F. Tesoriere A, et al. Biomedicines. 2021 Aug 4;9(8):956. doi: 10.3390/biomedicines9080956. Biomedicines. 2021. PMID: 34440160 Free PMC article. Review. - [Research progress of lung cancer on single nuleotide polymorphism].
Hua F, Zhou Q. Hua F, et al. Zhongguo Fei Ai Za Zhi. 2011 Feb;14(2):156-64. doi: 10.3779/j.issn.1009-3419.2011.02.10. Zhongguo Fei Ai Za Zhi. 2011. PMID: 21342648 Free PMC article. Review. Chinese. No abstract available.
Cited by
- PIAS3 expression in squamous cell lung cancer is low and predicts overall survival.
Abbas R, McColl KS, Kresak A, Yang M, Chen Y, Fu P, Wildey G, Dowlati A. Abbas R, et al. Cancer Med. 2015 Mar;4(3):325-32. doi: 10.1002/cam4.372. Epub 2015 Jan 9. Cancer Med. 2015. PMID: 25573684 Free PMC article. - Negative regulators of STAT3 signaling pathway in cancers.
Wu M, Song D, Li H, Yang Y, Ma X, Deng S, Ren C, Shu X. Wu M, et al. Cancer Manag Res. 2019 May 29;11:4957-4969. doi: 10.2147/CMAR.S206175. eCollection 2019. Cancer Manag Res. 2019. PMID: 31213912 Free PMC article. Review. - Therapeutic Potential of Targeting the SUMO Pathway in Cancer.
Kukkula A, Ojala VK, Mendez LM, Sistonen L, Elenius K, Sundvall M. Kukkula A, et al. Cancers (Basel). 2021 Aug 31;13(17):4402. doi: 10.3390/cancers13174402. Cancers (Basel). 2021. PMID: 34503213 Free PMC article. Review. - Multicellular Effects of STAT3 in Non-small Cell Lung Cancer: Mechanistic Insights and Therapeutic Opportunities.
Parakh S, Ernst M, Poh AR. Parakh S, et al. Cancers (Basel). 2021 Dec 11;13(24):6228. doi: 10.3390/cancers13246228. Cancers (Basel). 2021. PMID: 34944848 Free PMC article. Review. - Combining STAT3-Targeting Agents with Immune Checkpoint Inhibitors in NSCLC.
Papavassiliou KA, Marinos G, Papavassiliou AG. Papavassiliou KA, et al. Cancers (Basel). 2023 Jan 6;15(2):386. doi: 10.3390/cancers15020386. Cancers (Basel). 2023. PMID: 36672335 Free PMC article.
References
- Arriagada, R. , Bergman, B. , Dunant, A. , Le Chevalier, T. , Pignon, J.P. , Vansteenkiste, J. , 2004. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N. Engl. J. Med.. 350, 351–360. - PubMed
- Brantley, E.C. , Nabors, L.B. , Gillespie, G.Y. , Choi, Y.H. , Palmer, C.A. , Harrison, K. , Roarty, K. , Benveniste, E.N. , 2008. Loss of protein inhibitors of activated STAT-3 expression in glioblastoma multiforme tumors: implications for STAT-3 activation and gene expression. Clin. Cancer Res.. 14, 4694–4704. - PMC - PubMed
- Buettner, R. , Mora, L.B. , Jove, R. , 2002. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Cancer Res.. 8, 945–954. - PubMed
- Chung, C.D. , Liao, J. , Liu, B. , Rao, X. , Jay, P. , Berta, P. , Shuai, K. , 1997. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 278, 1803–1805. - PubMed
- Cortas, T. , Eisenberg, R. , Fu, P. , Kern, J. , Patrick, L. , Dowlati, A. , 2007. Activation state EGFR and STAT-3 as prognostic markers in resected non-small cell lung cancer. Lung Cancer. 55, 349–355. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous