Alphacoronaviruses in New World bats: prevalence, persistence, phylogeny, and potential for interaction with humans - PubMed (original) (raw)
Alphacoronaviruses in New World bats: prevalence, persistence, phylogeny, and potential for interaction with humans
Christina Osborne et al. PLoS One. 2011.
Abstract
Bats are reservoirs for many different coronaviruses (CoVs) as well as many other important zoonotic viruses. We sampled feces and/or anal swabs of 1,044 insectivorous bats of 2 families and 17 species from 21 different locations within Colorado from 2007 to 2009. We detected alphacoronavirus RNA in bats of 4 species: big brown bats (Eptesicus fuscus), 10% prevalence; long-legged bats (Myotis volans), 8% prevalence; little brown bats (Myotis lucifugus), 3% prevalence; and western long-eared bats (Myotis evotis), 2% prevalence. Overall, juvenile bats were twice as likely to be positive for CoV RNA as adult bats. At two of the rural sampling sites, CoV RNAs were detected in big brown and long-legged bats during the three sequential summers of this study. CoV RNA was detected in big brown bats in all five of the urban maternity roosts sampled throughout each of the periods tested. Individually tagged big brown bats that were positive for CoV RNA and later sampled again all became CoV RNA negative. Nucleotide sequences in the RdRp gene fell into 3 main clusters, all distinct from those of Old World bats. Similar nucleotide sequences were found in amplicons from gene 1b and the spike gene in both a big-brown and a long-legged bat, indicating that a CoV may be capable of infecting bats of different genera. These data suggest that ongoing evolution of CoVs in bats creates the possibility of a continued threat for emergence into hosts of other species. Alphacoronavirus RNA was detected at a high prevalence in big brown bats in roosts in close proximity to human habitations (10%) and known to have direct contact with people (19%), suggesting that significant potential opportunities exist for cross-species transmission of these viruses. Further CoV surveillance studies in bats throughout the Americas are warranted.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
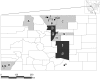
Figure 1. Map of Colorado showing sites where bats were sampled for the presence of CoV RNA.
Circles (#1–21) represent sites where live bats were captured and fecal or swab samples were taken; closed circles represent sites where bats tested positive for CoV RNA and open circles are those from which all samples tested negative. Shaded counties (A–K) were those from which intestines of bats submitted to public health departments were sampled for CoV RNA. Counties from which intestinal samples were negative for CoV are shown in gray and counties with at least one CoV-positive intestinal sample are shown in black.
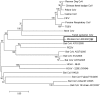
Figure 2. Phylogenetic Analysis of the spike gene.
Phylogenetic analysis of an 1100 nucleotide segment of the S2 region of the spike gene of RM-Bat-CoV 453/2007/EF (Eptesicus fuscus) compared to other known alphacoronaviruses. Phylogenetic trees were constructed by the neighbor-joining method using MEGA version 4.
Figure 3. Phylogenetic analysis of the RdRp gene.
Phylogenetic analysis of an approximate 4000 nucleotide sequence (2 segments) of the RdRp gene of RM-Bat-CoV-15/2006/ML (Myotis lucifugus) and RM-Bat-CoV 61/2007/EF (Eptesicus fuscus) compared to other known coronaviruses. Phylogenetic trees were constructed by the neighbor-joining method using MEGA version 4.
Similar articles
- Prevalence of neutralizing antibodies to rabies virus in serum of seven species of insectivorous bats from Colorado and New Mexico, United States.
Bowen RA, O'Shea TJ, Shankar V, Neubaum MA, Neubaum DJ, Rupprecht CE. Bowen RA, et al. J Wildl Dis. 2013 Apr;49(2):367-74. doi: 10.7589/2012-05-124. J Wildl Dis. 2013. PMID: 23568912 - Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.
Drexler JF, Corman VM, Drosten C. Drexler JF, et al. Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review. - Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China.
He B, Zhang Y, Xu L, Yang W, Yang F, Feng Y, Xia L, Zhou J, Zhen W, Feng Y, Guo H, Zhang H, Tu C. He B, et al. J Virol. 2014 Jun;88(12):7070-82. doi: 10.1128/JVI.00631-14. Epub 2014 Apr 9. J Virol. 2014. PMID: 24719429 Free PMC article. - Detection of new genetic variants of Betacoronaviruses in Endemic Frugivorous Bats of Madagascar.
Razanajatovo NH, Nomenjanahary LA, Wilkinson DA, Razafimanahaka JH, Goodman SM, Jenkins RK, Jones JP, Heraud JM. Razanajatovo NH, et al. Virol J. 2015 Mar 12;12:42. doi: 10.1186/s12985-015-0271-y. Virol J. 2015. PMID: 25888853 Free PMC article. - Bat origin of human coronaviruses.
Hu B, Ge X, Wang LF, Shi Z. Hu B, et al. Virol J. 2015 Dec 22;12:221. doi: 10.1186/s12985-015-0422-1. Virol J. 2015. PMID: 26689940 Free PMC article. Review.
Cited by
- Genetic diversity of bats coronaviruses in the Atlantic Forest hotspot biome, Brazil.
Góes LGB, Campos ACA, Carvalho C, Ambar G, Queiroz LH, Cruz-Neto AP, Munir M, Durigon EL. Góes LGB, et al. Infect Genet Evol. 2016 Oct;44:510-513. doi: 10.1016/j.meegid.2016.07.034. Epub 2016 Jul 26. Infect Genet Evol. 2016. PMID: 27473780 Free PMC article. - Novel bat coronaviruses, Brazil and Mexico.
Góes LG, Ruvalcaba SG, Campos AA, Queiroz LH, de Carvalho C, Jerez JA, Durigon EL, Dávalos LI, Dominguez SR. Góes LG, et al. Emerg Infect Dis. 2013 Oct;19(10):1711-3. doi: 10.3201/eid1910.130525. Emerg Infect Dis. 2013. PMID: 24050144 Free PMC article. No abstract available. - Viral maintenance and excretion dynamics of coronaviruses within an Egyptian rousette fruit bat maternal colony: considerations for spillover.
Geldenhuys M, Ross N, Dietrich M, de Vries JL, Mortlock M, Epstein JH, Weyer J, Pawęska JT, Markotter W. Geldenhuys M, et al. Sci Rep. 2023 Sep 22;13(1):15829. doi: 10.1038/s41598-023-42938-w. Sci Rep. 2023. PMID: 37739999 Free PMC article. - Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation.
Qian Z, Dominguez SR, Holmes KV. Qian Z, et al. PLoS One. 2013 Oct 3;8(10):e76469. doi: 10.1371/journal.pone.0076469. eCollection 2013. PLoS One. 2013. PMID: 24098509 Free PMC article. - Persistent infections support maintenance of a coronavirus in a population of Australian bats (Myotis macropus).
Jeong J, Smith CS, Peel AJ, Plowright RK, Kerlin DH, McBroom J, McCallum H. Jeong J, et al. Epidemiol Infect. 2017 Jul;145(10):2053-2061. doi: 10.1017/S0950268817000991. Epub 2017 May 22. Epidemiol Infect. 2017. PMID: 28528587 Free PMC article.
References
- Selvey LA, Wells RM, McCormack JG, Ansford AJ, Murray K, et al. Infection of humans and horses by a newly described morbillivirus. Med J Aust. 1995;162:642–645. - PubMed
Publication types
MeSH terms
Grants and funding
- 5K08-AI073525-03/AI/NIAID NIH HHS/United States
- U54-AI065357/AI/NIAID NIH HHS/United States
- K08 AI073525/AI/NIAID NIH HHS/United States
- U54 AI065357/AI/NIAID NIH HHS/United States
- AI-P01-059576/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials