Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo - PubMed (original) (raw)
Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo
Rodolphe Suspène et al. J Virol. 2011 Aug.
Abstract
Human APOBEC3 cytidine deaminases target and edit single-stranded DNA, which can be of viral, mitochondrial, or nuclear origin. Retrovirus genomes, such as human immunodeficiency virus (HIV) genomes deficient in the vif gene and the hepatitis B virus genome, are particularly vulnerable. The genomes of some DNA viruses, such as human papillomaviruses, can be edited in vivo and in transfection experiments. Accordingly, herpesviruses should be no exception. This is indeed the case for herpes simplex virus 1 (HSV-1) in tissue culture, where APOBEC3C (A3C) overexpression can reduce virus titers and the particle/PFU ratio ∼10-fold. Nonetheless, A3A, A3G, and AICDA can edit what is presumably a small fraction of HSV genomes in an experimental setting without seriously impacting the viral titer. Hyperediting was found in HSV genomes recovered from 4/8 uncultured buccal lesions. The phenomenon is not restricted to HSV, since hyperedited Epstein-Barr virus (EBV) genomes were readily recovered from 4/5 established cell lines, indicating that episomes are vulnerable to editing. These findings suggest that the widely expressed A3C cytidine deaminase can function as a restriction factor for some human herpesviruses. That the A3C gene is not induced by type I interferons begs the question whether some herpesviruses encode A3C antagonists.
Figures
Fig. 1.
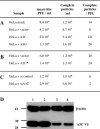
Several human A3 enzymes may restrict HSV-1 replication and edit the genome. (A) Transfection of HeLa cells by 2.5 μg of plasmid DNA followed by HSV-1 infection. pv, empty vector. Titers are the means for triplicate experiments. (B) Dose-response relationship of supernatant titers as a function of A3C DNA concentration. The DNA concentration was maintained constant with the plasmid vector. (C) 3DPCR of HSV-1 DNA for all DNA transfections. The denaturation temperature (_T_d) is given across the top; M, molecular weight markers. The HSV-1 sequence between the primers is 410 bp in length. The white vertical bar indicates the minimal _T_d (93 to 93.8°C) for cloned wild-type HSV-1 DNA. Bands to lower temperature can be taken as prima facie evidence of cytidine deamination. (D) A selection of A3-edited HSV-1 genomes from the HeLa + pv transfection. For clarity, only 200 bp of the 410-bp ICP22 sequence are shown, while differences are scored with respect to the reference sequence (G → A, n = 25, 10,250 bp; C → T, n = 9, 3,690 bp). The positions in the HSV-1 strain 17 sequence are also given. To the right of each sequence is the total number of C → T transitions on the plus or minus (shown as G → A transitions) strand.
Fig. 2.
A3C levels can impact particle/PFU ratios for HSV-1. (A) Impact of active A3 constructs. (B) Impact of the A3C C957S inactive mutant. (C) Impact of A3C siRNA. (D) Western blot of V5-tagged A3C and β-actin loading control for uninfected HeLa cells at 48 h; lanes 1 and 2, 400 ng A3C-V5 tag plus 1 μg siRNA control; lanes 3 and 4, 400 ng A3C-V5 tag plus 1 μg siA3C RNA. Molecular mass markers (in kDa) are shown to the left.
Fig. 3.
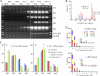
Four human cytidine deaminases can hyperedit HSV-1 genomes. (A) Agarose gels of ICP22 3DPCR DNA products from HSV-1-infected quail QT6 cells expressing various A3 genes. The annotation is as for Fig. 1C. Stars indicate A3A, A3C, and A3G catalytic mutants. (B) 5′ dinucleotide context associated with editing on the plus and minus strands. Asterisks denote significant deviations from the expected values (χ2 test, P < 0.05) (QT6 + A3A, G → A, n = 24, 9,840 bp; C → T, n = 31, 12,710 bp; QT6 + A3C, G → A, n = 50, 20,500 bp; C → T, n = 78, 31,980 bp; QT6 + A3G, G → A, n = 32, 13,120 bp; C → T, n = 8, 3,280 bp; QT6 + AID, G → A, n = 9, 3,690 bp; C → T, n = 24, 9,840 bp). (C) TaqMan transcriptome analysis of the seven A3 genes from HeLa cells. The levels have been normalized to those of A3G (horizontal bar). nd, not detected.
Fig. 4.
HSV-1 genomes can be edited in vivo. (A) Agarose gels of ICP22 3DPCR DNA products from total DNA derived from pharyngeal washes or labial swabs. The annotation is as for Fig. 1C. (B) Frequency distribution of A3-edited HSV-1 sequences from patient P9. Since the cytidine composition of the two strands varies (32% plus strand; 35% minus strand), the % editing differs slightly (P9 G → A, n = 88, 36,080 bp; C → T, n = 21, 8,610 bp). (C) 5′ dinucleotide context associated with editing on the plus and minus strands for the collection of sequences from patient P9 (_T_d = 90.4 to 92.1°C). Asterisks denote significant deviations form the expected values (χ2 test, P < 0.05). Also shown are sequences derived at a higher _T_d (93.8 to 94.5°C) with 1 to 4 C → T transitions or 5 to 10 transitions pooled from all patients [Patients (1–4 mut.), n = 713, 292,330 bp; Patients (5–10 mut.), n = 10, 4,100 bp; P9, G → A, n = 94, 38,540 bp; C → T, n = 21, 8,610 bp]. (D) Frequency distribution of sequences derived at 93.8 to 94.5°C from all eight patients for both strands. The insets expand the region covering 4 to 55 bp.
Fig. 5.
Hyperediting of EBV genomes in EBV-immortalized human B cell lines. (A) Agarose gels of EBNA-1 3DPCR DNA products from total DNA derived from five EBV-immortalized human B cell lines. The annotation is as for Fig. 1C. The P1 to -3 lines are all _ung_−/−; Z is _AICDA_−/−, while T is an EBV-cell line from a normal patient without any known leukocyte defect. (B) A selection of hyperedited sequences from P1 to -3 and Z. To the right are the number and % of cytidine residues edited. Plus-strand hypermutants were not always recovered and hence were excluded from analyses. (C) Frequency distribution of A3-edited EBV sequences from four EBV-transformed cell lines. Only minus-strand editing was identified (P1, n = 32, 6,400 bp; P2, n = 21, 4,200 bp; P3, n = 19, 3,800 bp; Z, n = 72, 14,400 bp). (D) 5′ dinucleotide context associated with minus-strand editing. Asterisks denote significant deviations from the expected values (χ2 test, P < 0.05).
Similar articles
- A Conserved Mechanism of APOBEC3 Relocalization by Herpesviral Ribonucleotide Reductase Large Subunits.
Cheng AZ, Moraes SN, Attarian C, Yockteng-Melgar J, Jarvis MC, Biolatti M, Galitska G, Dell'Oste V, Frappier L, Bierle CJ, Rice SA, Harris RS. Cheng AZ, et al. J Virol. 2019 Nov 13;93(23):e01539-19. doi: 10.1128/JVI.01539-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31534038 Free PMC article. - Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis.
Vartanian JP, Henry M, Marchio A, Suspène R, Aynaud MM, Guétard D, Cervantes-Gonzalez M, Battiston C, Mazzaferro V, Pineau P, Dejean A, Wain-Hobson S. Vartanian JP, et al. PLoS Pathog. 2010 May 27;6(5):e1000928. doi: 10.1371/journal.ppat.1000928. PLoS Pathog. 2010. PMID: 20523896 Free PMC article. - Host restriction of murine gammaherpesvirus 68 replication by human APOBEC3 cytidine deaminases but not murine APOBEC3.
Minkah N, Chavez K, Shah P, Maccarthy T, Chen H, Landau N, Krug LT. Minkah N, et al. Virology. 2014 Apr;454-455:215-26. doi: 10.1016/j.virol.2014.02.022. Epub 2014 Mar 13. Virology. 2014. PMID: 24725948 Free PMC article. - Hepatitis B: modern concepts in pathogenesis--APOBEC3 cytidine deaminases as effectors in innate immunity against the hepatitis B virus.
Bonvin M, Greeve J. Bonvin M, et al. Curr Opin Infect Dis. 2008 Jun;21(3):298-303. doi: 10.1097/QCO.0b013e3282fe1bb2. Curr Opin Infect Dis. 2008. PMID: 18448976 Review. - Herpes simplex virus type 1 amplicons and their hybrid virus partners, EBV, AAV, and retrovirus.
Oehmig A, Fraefel C, Breakefield XO, Ackermann M. Oehmig A, et al. Curr Gene Ther. 2004 Dec;4(4):385-408. doi: 10.2174/1566523043346129. Curr Gene Ther. 2004. PMID: 15578989 Review.
Cited by
- Efficient deamination of 5-methylcytidine and 5-substituted cytidine residues in DNA by human APOBEC3A cytidine deaminase.
Suspène R, Aynaud MM, Vartanian JP, Wain-Hobson S. Suspène R, et al. PLoS One. 2013 Jun 20;8(6):e63461. doi: 10.1371/journal.pone.0063461. Print 2013. PLoS One. 2013. PMID: 23840298 Free PMC article. - A Multi-Laboratory Comparison of Methods for Detection and Quantification of African Swine Fever Virus.
Olesen AS, Bruun Rasmussen T, Saxmose Nielsen S, Belsham GJ, Boklund A, Ploegaert T, Moonen-Leusen B, Blome S, Bøtner A. Olesen AS, et al. Pathogens. 2022 Mar 7;11(3):325. doi: 10.3390/pathogens11030325. Pathogens. 2022. PMID: 35335649 Free PMC article. - Human cytomegalovirus mediates APOBEC3B relocalization early during infection through a ribonucleotide reductase-independent mechanism.
Fanunza E, Cheng AZ, Auerbach AA, Stefanovska B, Moraes SN, Lokensgard JR, Biolatti M, Dell'Oste V, Bierle CJ, Bresnahan WA, Harris RS. Fanunza E, et al. bioRxiv [Preprint]. 2023 Jan 31:2023.01.30.526383. doi: 10.1101/2023.01.30.526383. bioRxiv. 2023. PMID: 36778493 Free PMC article. Updated. Preprint. - Human cytomegalovirus mediates APOBEC3B relocalization early during infection through a ribonucleotide reductase-independent mechanism.
Fanunza E, Cheng AZ, Auerbach AA, Stefanovska B, Moraes SN, Lokensgard JR, Biolatti M, Dell'Oste V, Bierle CJ, Bresnahan WA, Harris RS. Fanunza E, et al. J Virol. 2023 Aug 31;97(8):e0078123. doi: 10.1128/jvi.00781-23. Epub 2023 Aug 11. J Virol. 2023. PMID: 37565748 Free PMC article. - Evolutionary effects of the AID/APOBEC family of mutagenic enzymes on human gamma-herpesviruses.
Martinez T, Shapiro M, Bhaduri-McIntosh S, MacCarthy T. Martinez T, et al. Virus Evol. 2019 Feb 11;5(1):vey040. doi: 10.1093/ve/vey040. eCollection 2019 Jan. Virus Evol. 2019. PMID: 30792902 Free PMC article.
References
- Beale R. C., et al. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585–596 - PubMed
- Bishop K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396 - PubMed
- Bonvin M., et al. 2006. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43:1364–1374 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases