Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia - PubMed (original) (raw)
. 2011 Jun 5;475(7354):101-5.
doi: 10.1038/nature10113.
Magda Pinyol, Víctor Quesada, Laura Conde, Gonzalo R Ordóñez, Neus Villamor, Georgia Escaramis, Pedro Jares, Sílvia Beà, Marcos González-Díaz, Laia Bassaganyas, Tycho Baumann, Manel Juan, Mónica López-Guerra, Dolors Colomer, José M C Tubío, Cristina López, Alba Navarro, Cristian Tornador, Marta Aymerich, María Rozman, Jesús M Hernández, Diana A Puente, José M P Freije, Gloria Velasco, Ana Gutiérrez-Fernández, Dolors Costa, Anna Carrió, Sara Guijarro, Anna Enjuanes, Lluís Hernández, Jordi Yagüe, Pilar Nicolás, Carlos M Romeo-Casabona, Heinz Himmelbauer, Ester Castillo, Juliane C Dohm, Silvia de Sanjosé, Miguel A Piris, Enrique de Alava, Jesús San Miguel, Romina Royo, Josep L Gelpí, David Torrents, Modesto Orozco, David G Pisano, Alfonso Valencia, Roderic Guigó, Mónica Bayés, Simon Heath, Marta Gut, Peter Klatt, John Marshall, Keiran Raine, Lucy A Stebbings, P Andrew Futreal, Michael R Stratton, Peter J Campbell, Ivo Gut, Armando López-Guillermo, Xavier Estivill, Emili Montserrat, Carlos López-Otín, Elías Campo
Affiliations
- PMID: 21642962
- PMCID: PMC3322590
- DOI: 10.1038/nature10113
Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia
Xose S Puente et al. Nature. 2011.
Abstract
Chronic lymphocytic leukaemia (CLL), the most frequent leukaemia in adults in Western countries, is a heterogeneous disease with variable clinical presentation and evolution. Two major molecular subtypes can be distinguished, characterized respectively by a high or low number of somatic hypermutations in the variable region of immunoglobulin genes. The molecular changes leading to the pathogenesis of the disease are still poorly understood. Here we performed whole-genome sequencing of four cases of CLL and identified 46 somatic mutations that potentially affect gene function. Further analysis of these mutations in 363 patients with CLL identified four genes that are recurrently mutated: notch 1 (NOTCH1), exportin 1 (XPO1), myeloid differentiation primary response gene 88 (MYD88) and kelch-like 6 (KLHL6). Mutations in MYD88 and KLHL6 are predominant in cases of CLL with mutated immunoglobulin genes, whereas NOTCH1 and XPO1 mutations are mainly detected in patients with unmutated immunoglobulins. The patterns of somatic mutation, supported by functional and clinical analyses, strongly indicate that the recurrent NOTCH1, MYD88 and XPO1 mutations are oncogenic changes that contribute to the clinical evolution of the disease. To our knowledge, this is the first comprehensive analysis of CLL combining whole-genome sequencing with clinical characteristics and clinical outcomes. It highlights the usefulness of this approach for the identification of clinically relevant mutations in cancer.
©2011 Macmillan Publishers Limited. All rights reserved
Figures
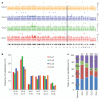
Figure 1. Profile of somatic mutations in four CLL genomes
a, Distribution of somatic alterations. For each tumour genome, copy number (solid lines), density of mutations per 5-Mb window (bars) and protein-coding mutations (dots) are shown. The shaded rectangle indicates the location of the 13q14 deletion that was present in three of the four CLL cases. Chromosome numbers are listed below the four profiles. b, Frequency of substitutions in each CLL tumour for the six possible classes of mutation. c, Distribution of the four possible NpA dinucleotides for the A to C transversion in each tumour genome, compared with the expected distribution across the genome. The total number of A to C substitutions per case is indicated at the top (**, P < 0.001).
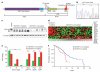
Figure 2. Mutational and functional analysis of NOTCH1 in CLL
a, Schematic representation of human NOTCH1, showing the main domains and locations of the three different somatic mutations identified in CLL. NEC, NOTCH1 extracellular subunit; NTM, NOTCH1 transmembrane subunit; ICN, intracellular domain of NOTCH1; LNR, Lin-12 NOTCH repeats; RAM, RAM domain; ANK, ankyrin repeat domain; PEST, PEST domain. b, Electropherogram showing the heterozygous CT deletion recurrently identified in CLL. c, Western blot showing NOTCH1 protein levels in CLL cases with or without the NOTCH1 p.P2515Rfs*4 mutation, and in Jurkat cells as a control. The arrow indicates the band corresponding to the NTM; the large arrowhead indicates the smaller band corresponding to the mutant form. d, Heat map showing the 23 genes of the NOTCH1 pathway that are differentially expressed in _NOTCH1_-mutated versus non-mutated CLL. e, Distribution of disease stage (Binet), ZAP-70 expression status, CD38 expression status and IGHV mutational status (UM, unmutated IGHV) in patients with or without mutations in NOTCH1 (*, P < 0.02; **, P < 0.01). f, Actuarial probability of overall survival of CLL patients with mutated or unmutated NOTCH1 (*, P = 0.03).
Figure 3. Mutational and functional analysis of MYD88 in CLL
a, Multiple sequence alignment of MyD88 around the mutated residue (arrow) in different species. Cons., degree of conservation. b, Electropherogram showing the recurrent heterozygous p.L265P MYD88 mutation (arrow) detected in CLL. c, Cell extracts from a _MYD88_-mutated CLL (L265P) and a _MYD88_-unmutated CLL (WT) were immunoprecipitated with anti-MyD88 antibody. The immunoprecipitated and unbound fractions were analysed by western blot using anti-IRAK1 and anti-MyD88 antibodies. d, Western blot analysis of phosphorylated STAT3 (p-STAT3 (Tyr 705)) and total STAT3 in cell extracts from _MYD88_-mutated or unmutated CLL tumour cells. β-Actin was used as a control to show equal loading. e, Western blots showing phosphorylated IκBα (p-IκBα), total IκBα, phosphorylated p65 subunit of NF-κB (p-p65) and total p65 subunit of NF-κB in cell extracts from _MYD88_-mutated or unmutated CLL tumour cells. f, Heat map representing the cytokine levels secreted by B cells from eight individuals after Toll-like receptor stimulation. Only the five cytokines that showed the most significant differences between _MYD88_-mutated and _MYD88_-unmutated CLL are shown. ‘E52DEL’ indicates B cells from two patients with an inactivating MYD88 mutation, ‘WT’ corresponds to tumour cells from CLL patients without MYD88 mutation and ‘L265P’ indicates tumour cells from patients carrying a mutated MYD88. The stimulation experiments for each of the Toll-like receptors (TLRs) are represented in different colours. NS, no stimulus. g, Distribution of disease stage (Binet), ZAP-70 expression status, CD38 expression status and IGHV mutational status (UM, unmutated IGHV) in patients according to the presence or absence of p.L265P MYD88 mutation (*, P < 0.03).
Comment in
- Genetics: Clues for targeting leukemia.
Schmitt CA. Schmitt CA. Nat Rev Clin Oncol. 2011 Jun 21;8(8):447. doi: 10.1038/nrclinonc.2011.96. Nat Rev Clin Oncol. 2011. PMID: 21691320 No abstract available. - Comprehensive identification of somatic mutations in chronic lymphocytic leukemia.
Bergsagel PL, Kuehl WM. Bergsagel PL, et al. Cancer Cell. 2011 Jul 12;20(1):5-7. doi: 10.1016/j.ccr.2011.06.023. Cancer Cell. 2011. PMID: 21741593 Free PMC article.
Similar articles
- A reduced panel of eight genes (ATM, SF3B1, NOTCH1, BIRC3, XPO1, MYD88, TNFAIP3, and TP53) as an estimator of the tumor mutational burden in chronic lymphocytic leukemia.
Chauzeix J, Pastoret C, Donaty L, Gachard N, Fest T, Feuillard J, Rizzo D. Chauzeix J, et al. Int J Lab Hematol. 2021 Aug;43(4):683-692. doi: 10.1111/ijlh.13435. Epub 2020 Dec 16. Int J Lab Hematol. 2021. PMID: 33325634 Free PMC article. - NOTCH1 mutations in CLL associated with trisomy 12.
Balatti V, Bottoni A, Palamarchuk A, Alder H, Rassenti LZ, Kipps TJ, Pekarsky Y, Croce CM. Balatti V, et al. Blood. 2012 Jan 12;119(2):329-31. doi: 10.1182/blood-2011-10-386144. Epub 2011 Nov 15. Blood. 2012. PMID: 22086416 Free PMC article. - SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients.
Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V, Alpermann T, Roller A, Kohlmann A, Haferlach T, Kern W, Schnittger S. Jeromin S, et al. Leukemia. 2014 Jan;28(1):108-17. doi: 10.1038/leu.2013.263. Epub 2013 Sep 12. Leukemia. 2014. PMID: 24113472 - Recurrent gene mutations in CLL.
Martínez-Trillos A, Quesada V, Villamor N, Puente XS, López-Otín C, Campo E. Martínez-Trillos A, et al. Adv Exp Med Biol. 2013;792:87-107. doi: 10.1007/978-1-4614-8051-8_4. Adv Exp Med Biol. 2013. PMID: 24014293 Review. - Overview of non-coding mutations in chronic lymphocytic leukemia.
Spina V, Rossi D. Spina V, et al. Mol Oncol. 2019 Jan;13(1):99-106. doi: 10.1002/1878-0261.12416. Epub 2019 Jan 4. Mol Oncol. 2019. PMID: 30520556 Free PMC article. Review.
Cited by
- Genomic sequencing in cancer.
Tuna M, Amos CI. Tuna M, et al. Cancer Lett. 2013 Nov 1;340(2):161-70. doi: 10.1016/j.canlet.2012.11.004. Epub 2012 Nov 23. Cancer Lett. 2013. PMID: 23178448 Free PMC article. Review. - MiR-181b: new perspective to evaluate disease progression in chronic lymphocytic leukemia.
Visone R, Veronese A, Balatti V, Croce CM. Visone R, et al. Oncotarget. 2012 Feb;3(2):195-202. doi: 10.18632/oncotarget.448. Oncotarget. 2012. PMID: 22350310 Free PMC article. - Exome sequencing of early-onset patients supports genetic heterogeneity in colorectal cancer.
Fernández-Rozadilla C, Álvarez-Barona M, Quintana I, López-Novo A, Amigo J, Cameselle-Teijeiro JM, Roman E, Gonzalez D, Llor X, Bujanda L, Bessa X, Jover R, Balaguer F, Castells A, Castellví-Bel S, Capellá G, Carracedo A, Valle L, Ruiz-Ponte C. Fernández-Rozadilla C, et al. Sci Rep. 2021 May 27;11(1):11135. doi: 10.1038/s41598-021-90590-z. Sci Rep. 2021. PMID: 34045552 Free PMC article. - Targeting ATM-deficient CLL through interference with DNA repair pathways.
Knittel G, Liedgens P, Reinhardt HC. Knittel G, et al. Front Genet. 2015 Jun 10;6:207. doi: 10.3389/fgene.2015.00207. eCollection 2015. Front Genet. 2015. PMID: 26113859 Free PMC article. Review. - Phase 1 study of 2 high dose intensity schedules of the pan-Notch inhibitor crenigacestat (LY3039478) in combination with prednisone in patients with advanced or metastatic cancer.
Azaro A, Baldini C, Rodon J, Soria JC, Yuen E, Lithio A, Oakley G, Benhadji KA, Massard C. Azaro A, et al. Invest New Drugs. 2021 Feb;39(1):193-201. doi: 10.1007/s10637-020-00944-z. Epub 2020 Sep 11. Invest New Drugs. 2021. PMID: 32915419 Clinical Trial.
References
- Rozman C, Montserrat E. Chronic lymphocytic leukemia. N. Engl. J. Med. 1995;333:1052–1057. - PubMed
- Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nature Rev. Cancer. 2010;10:37–50. - PubMed
- Damle RN, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. - PubMed
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases