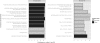g:Profiler--a web server for functional interpretation of gene lists (2011 update) - PubMed (original) (raw)
. 2011 Jul;39(Web Server issue):W307-15.
doi: 10.1093/nar/gkr378. Epub 2011 Jun 6.
Affiliations
- PMID: 21646343
- PMCID: PMC3125778
- DOI: 10.1093/nar/gkr378
g:Profiler--a web server for functional interpretation of gene lists (2011 update)
Jüri Reimand et al. Nucleic Acids Res. 2011 Jul.
Abstract
Functional interpretation of candidate gene lists is an essential task in modern biomedical research. Here, we present the 2011 update of g:Profiler (http://biit.cs.ut.ee/gprofiler/), a popular collection of web tools for functional analysis. g:GOSt and g:Cocoa combine comprehensive methods for interpreting gene lists, ordered lists and list collections in the context of biomedical ontologies, pathways, transcription factor and microRNA regulatory motifs and protein-protein interactions. Additional tools, namely the biomolecule ID mapping service (g:Convert), gene expression similarity searcher (g:Sorter) and gene homology searcher (g:Orth) provide numerous ways for further analysis and interpretation. In this update, we have implemented several features of interest to the community: (i) functional analysis of single nucleotide polymorphisms and other DNA polymorphisms is supported by chromosomal queries; (ii) network analysis identifies enriched protein-protein interaction modules in gene lists; (iii) functional analysis covers human disease genes; and (iv) improved statistics and filtering provide more concise results. g:Profiler is a regularly updated resource that is available for a wide range of species, including mammals, plants, fungi and insects.
Figures
Figure 1.
g:Profiler tools (A–E) and data streams (arrows) for constructing analysis pipelines. (A) g:GOSt—functional profiling of gene lists, with sources of input shown on the left [also see (F)]. Output: genes are shown horizontally and annotations vertically, colours denote classes of functional evidence. (B) g:Cocoa—functional profiling of multiple gene lists. Output: gene lists are shown horizontally and annotations vertically, colours (intensity of red) denote strength of enrichment. (C) g:Convert—ID mapping service for kinds of molecules and databases. (D) g:Sorter—gene-based expression similarity search from microarrays. (E) g:Orth—mapping homologous genes of related species. (F) Identification of enriched protein–protein interaction modules in gene lists. A fragment of g:GOSt output includes a module of interacting query genes (core, red) and non-query genes interacting with the core module (neighbourhood, black).
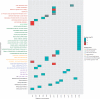
Figure 2.
Functional analysis of positively selected regions in modern human genome versus Neanderthal genome. Input genomic loci were grouped by chromosome and analysed with g:Cocoa. R was used for visualization. Enriched annotations are shown vertically and chromosomes horizontally. Coloured cells indicate statistically significant enrichments of corresponding functions (P < 0.05, red tones represent greater significance). Annotation axis labels are grouped and coloured according to data source. To avoid redundant annotations, only most significant function of any hierarchically related group is shown.
Figure 3.
Comparison of standard, global background gene list (left plot) and customized list (right plot) in functional enrichment analysis of nine core cell cycle transcription factors in yeast. Top 10 enriched categories are shown vertically and log-scale _P_-values horizontally. R was used for visualization. The global analysis reveals general functional enrichments of transcriptional regulation (black bars), while focused analysis with the custom background of all yeast TFs shows specific cell cycle-related terms (grey bars).
Similar articles
- g:Profiler-a web server for functional interpretation of gene lists (2016 update).
Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J. Reimand J, et al. Nucleic Acids Res. 2016 Jul 8;44(W1):W83-9. doi: 10.1093/nar/gkw199. Epub 2016 Apr 20. Nucleic Acids Res. 2016. PMID: 27098042 Free PMC article. - g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments.
Reimand J, Kull M, Peterson H, Hansen J, Vilo J. Reimand J, et al. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W193-200. doi: 10.1093/nar/gkm226. Epub 2007 May 3. Nucleic Acids Res. 2007. PMID: 17478515 Free PMC article. - g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update).
Kolberg L, Raudvere U, Kuzmin I, Adler P, Vilo J, Peterson H. Kolberg L, et al. Nucleic Acids Res. 2023 Jul 5;51(W1):W207-W212. doi: 10.1093/nar/gkad347. Nucleic Acids Res. 2023. PMID: 37144459 Free PMC article. - g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update).
Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. Raudvere U, et al. Nucleic Acids Res. 2019 Jul 2;47(W1):W191-W198. doi: 10.1093/nar/gkz369. Nucleic Acids Res. 2019. PMID: 31066453 Free PMC article. - A brief survey of tools for genomic regions enrichment analysis.
Chicco D, Jurman G. Chicco D, et al. Front Bioinform. 2022 Oct 26;2:968327. doi: 10.3389/fbinf.2022.968327. eCollection 2022. Front Bioinform. 2022. PMID: 36388843 Free PMC article. Review.
Cited by
- Biosignatures for Parkinson's disease and atypical parkinsonian disorders patients.
Potashkin JA, Santiago JA, Ravina BM, Watts A, Leontovich AA. Potashkin JA, et al. PLoS One. 2012;7(8):e43595. doi: 10.1371/journal.pone.0043595. Epub 2012 Aug 27. PLoS One. 2012. PMID: 22952715 Free PMC article. - Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity.
Meyer M, Reimand J, Lan X, Head R, Zhu X, Kushida M, Bayani J, Pressey JC, Lionel AC, Clarke ID, Cusimano M, Squire JA, Scherer SW, Bernstein M, Woodin MA, Bader GD, Dirks PB. Meyer M, et al. Proc Natl Acad Sci U S A. 2015 Jan 20;112(3):851-6. doi: 10.1073/pnas.1320611111. Epub 2015 Jan 5. Proc Natl Acad Sci U S A. 2015. PMID: 25561528 Free PMC article. - Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis.
Hinchcliff M, Huang CC, Wood TA, Matthew Mahoney J, Martyanov V, Bhattacharyya S, Tamaki Z, Lee J, Carns M, Podlusky S, Sirajuddin A, Shah SJ, Chang RW, Lafyatis R, Varga J, Whitfield ML. Hinchcliff M, et al. J Invest Dermatol. 2013 Aug;133(8):1979-89. doi: 10.1038/jid.2013.130. Epub 2013 Mar 14. J Invest Dermatol. 2013. PMID: 23677167 Free PMC article. - Differentiated neuroprogenitor cells incubated with human or canine adenovirus, or lentiviral vectors have distinct transcriptome profiles.
Piersanti S, Astrologo L, Licursi V, Costa R, Roncaglia E, Gennetier A, Ibanes S, Chillon M, Negri R, Tagliafico E, Kremer EJ, Saggio I. Piersanti S, et al. PLoS One. 2013 Jul 26;8(7):e69808. doi: 10.1371/journal.pone.0069808. Print 2013. PLoS One. 2013. PMID: 23922808 Free PMC article. - Methotrexate-induced neurotoxicity and leukoencephalopathy in childhood acute lymphoblastic leukemia.
Bhojwani D, Sabin ND, Pei D, Yang JJ, Khan RB, Panetta JC, Krull KR, Inaba H, Rubnitz JE, Metzger ML, Howard SC, Ribeiro RC, Cheng C, Reddick WE, Jeha S, Sandlund JT, Evans WE, Pui CH, Relling MV. Bhojwani D, et al. J Clin Oncol. 2014 Mar 20;32(9):949-59. doi: 10.1200/JCO.2013.53.0808. Epub 2014 Feb 18. J Clin Oncol. 2014. PMID: 24550419 Free PMC article.
References
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. - PubMed
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases