Resveratrol selectively induces DNA Damage, independent of Smad4 expression, in its efficacy against human head and neck squamous cell carcinoma - PubMed (original) (raw)
Resveratrol selectively induces DNA Damage, independent of Smad4 expression, in its efficacy against human head and neck squamous cell carcinoma
Alpna Tyagi et al. Clin Cancer Res. 2011.
Abstract
Purpose: Alterations in Smad4 signaling and its loss cause genomic instability and head and neck squamous cell carcinoma (HNSCC), suggesting that agents that target both Smad4-dependent and -independent pathways could control HNSCC.
Experimental design: Resveratrol efficacy was evaluated against the HNSCC cells FaDu, Cal27, Det562, and Cal27-Smad4 for viability, DNA damage, cell-cycle progression, and apoptosis, as well as γ-H2AX expression, and focus formation (γ-H2AX and Brca1). Resveratrol efficacy was also examined in nude mice for FaDu xenograft growth. Xenografts were analyzed for γ-H2AX and cleaved caspase-3.
Results: Resveratrol (5-50 μmol/L) suppressed viability and induced DNA damage in FaDu and Cal27 cells but not in normal human epidermal keratinocytes and human foreskin fibroblasts, showing its selectivity toward HNSCC cells; however, Det562 cells were resistant to resveratrol even at 100 μmol/L. Cal27 cells stably transfected with Smad4 showed similar resveratrol effects as parental Cal27, indicating that a lack of resveratrol effect in Det562 cells was independent of Smad4 status in these cells. Furthermore, resveratrol caused S-phase arrest and apoptotic death of FaDu and Cal27 cells together with induction of Brca1 and γ-H2AX foci. Resveratrol (50 mg/kg body weight) treatment also inhibited FaDu tumor growth in nude mice, and γ-H2AX and cleaved caspase-3 were strongly increased in xenografts from resveratrol-treated mice compared with controls.
Conclusion: Our findings for the first time showed antiproliferative, DNA damaging, and apoptotic effects of resveratrol in HNSCC cells independent of Smad4 status, both in vitro and in vivo, suggesting that more studies are needed to establish its potential usefulness against HNSCC.
©2011 AACR.
Figures
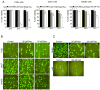
Figure 1. Resveratrol inhibits cell viability and induces DNA damage selectively in HNSCC cells
A) Resveratrol effect on cell viability of FaDu, Cal27 and Det562 cells employing MTT assay as detailed in Methods. *; p<0.001 between control and resveratrol; each bar represents mean±SE of three samples from each group. B) and C) Cells were treated for 24 h with indicated concentrations of resveratrol and then comet assay was performed as detailed in Methods. Representative pictures of damaged and undamaged nuclei in HNSCC FaDu, Cal27, Det562 cells (B), and NHEK and HFF cells (C) were captured as detailed in Methods. Res, resveratrol.
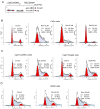
Figure 2. Resveratrol selectively causes cell cycle arrest in HNSCC cells independent of Smad4
A) Cal27 cells were transfected with pcDNA Flag-Smad4M (Plasmid 14959) or vector control pcDNA3 (Addgene, Cambridge, MA) using Lipofectamine 2000. Stable transfectants were selected using G418 at 0.2 mg/ml in DMEM (10% FBS) for 4 weeks, and cells were pooled and maintained in the same selective medium. Cells lysates were prepared and subjected to SDS-PAGE followed by immunoblotting, and membranes probed with Smad4 antibody as detailed in Methods. Percentage cell cycle distribution in B) FaDu, C) Cal27-pcDNA3 and Cal27-Smad4, and D) NHEK cells following resveratrol treatment for 24 h at indicated concentrations and FACS analysis as detailed in Methods. Results shown are representative of triplicate samples from each treatment, and were reproducible in two independent experiments. NS, nonspecific; Res, resveratrol.
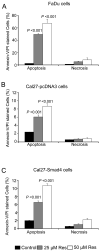
Figure 3. Resveratrol causes apoptotic death in HNSCC cells independent of Smad4
Percentage Annexin-V/PI stained apoptotic A) FaDu, B) Cal27-pcDNA3, and C) Cal27-Smad4 cells following resveratrol treatment for 48 h at indicated concentrations and flow cytometry analysis as detailed in Methods. Each bar represents the mean ± SE of the percentage of apoptotic cells from triplicate samples from each treatment, and these results were reproducible in three independent experiments. Res, resveratrol
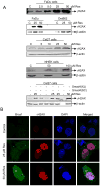
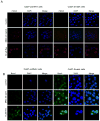
Figure 4. Resveratrol induces γH2AX and Brca1 foci formation in HNSCC cells
A) Cells were treated with DMSO alone or resveratrol for 24 h, total cell lysates prepared and subjected to SDS-PAGE followed by immunoblotting, and membranes probed with γH2AX and β-actin antibodies as detailed in Methods. B) Resveratrol induces the γH2AX and Brca1 foci formation. FaDu cells were treated with DMSO (control) or resveratrol (25-50 μM) for 24 h followed by immunocytochemical staining for γH2AX and Brca1 using specific antibody, cell images were captured at 1000× magnification on a Nikon D Eclipse C1 confocal microscope, and images analyzed by EZ-C1 Free viewer software. Red and green fluorescence represents staining for γH2AX and Brca1, respectively. Res, resveratrol; C, control.
Figure 5. Resveratrol-induced γH2AX and Brca1 foci formation is independent of Smad4 in Cal27 cells
Resveratrol induces A) γH2AX and B) Brca1 foci formation in Cal27-pcDNA3 and Cal27-Smad4 cells. Following indicated treatments of the cells, immunocytochemical staining for γH2AX and Brca1 was performed using specific antibody as detailed in Methods. Cell images were captured at 1000× magnification on a Nikon D Eclipse C1 confocal microscope, and analyzed by EZ-C1 Free viewer software. Red and green fluorescence represents staining for γH2AX and Brca1, respectively. Res, resveratrol.
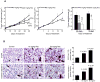
Figure 6. Resveratrol inhibits FaDu tumor xenograft growth together with in vivo DNA-damage and apoptotic effects
A) Growth inhibitory effects of resveratrol in terms of tumor volume as a function of treatment and tumor weight at study end following the experiment detailed in Methods. In each case, data shown are mean±SE of eight mice/group. B) IHC analyses for γH2AX and cleaved caspase-3 in FaDu tumor xenografts from the 30 days of resveratrol treatment study as detailed in Methods. Representative images are shown for both staining in all three groups' samples. In each case quantitative data are mean ± SE of five tumor samples from five individual mouse in each group from five randomly selected microscopic (400×) fields from each tumor. NS, not statistically significant; Res, resveratrol.
Similar articles
- Generation of reactive oxygen species by grape seed extract causes irreparable DNA damage leading to G2/M arrest and apoptosis selectively in head and neck squamous cell carcinoma cells.
Shrotriya S, Deep G, Gu M, Kaur M, Jain AK, Inturi S, Agarwal R, Agarwal C. Shrotriya S, et al. Carcinogenesis. 2012 Apr;33(4):848-58. doi: 10.1093/carcin/bgs019. Epub 2012 Jan 19. Carcinogenesis. 2012. PMID: 22266465 Free PMC article. - Mutations of the LIM protein AJUBA mediate sensitivity of head and neck squamous cell carcinoma to treatment with cell-cycle inhibitors.
Zhang M, Singh R, Peng S, Mazumdar T, Sambandam V, Shen L, Tong P, Li L, Kalu NN, Pickering CR, Frederick M, Myers JN, Wang J, Johnson FM. Zhang M, et al. Cancer Lett. 2017 Apr 28;392:71-82. doi: 10.1016/j.canlet.2017.01.024. Epub 2017 Jan 23. Cancer Lett. 2017. PMID: 28126323 Free PMC article. - PARP Inhibition Enhances Radiotherapy of SMAD4-Deficient Human Head and Neck Squamous Cell Carcinomas in Experimental Models.
Hernandez AL, Young CD, Bian L, Weigel K, Nolan K, Frederick B, Han G, He G, Devon Trahan G, Rudolph MC, Jones KL, Oweida AJ, Karam SD, Raben D, Wang XJ. Hernandez AL, et al. Clin Cancer Res. 2020 Jun 15;26(12):3058-3070. doi: 10.1158/1078-0432.CCR-19-0514. Epub 2020 Mar 5. Clin Cancer Res. 2020. PMID: 32139402 Free PMC article. - Emerging Phytochemicals for the Prevention and Treatment of Head and Neck Cancer.
Katiyar SK. Katiyar SK. Molecules. 2016 Nov 24;21(12):1610. doi: 10.3390/molecules21121610. Molecules. 2016. PMID: 27886147 Free PMC article. Review. - Two sides of the story? Smad4 loss in pancreatic cancer versus head-and-neck cancer.
Malkoski SP, Wang XJ. Malkoski SP, et al. FEBS Lett. 2012 Jul 4;586(14):1984-92. doi: 10.1016/j.febslet.2012.01.054. Epub 2012 Feb 3. FEBS Lett. 2012. PMID: 22321641 Free PMC article. Review.
Cited by
- The Potential of Phytochemicals in Oral Cancer Prevention and Therapy: A Review of the Evidence.
Lee TY, Tseng YH. Lee TY, et al. Biomolecules. 2020 Aug 6;10(8):1150. doi: 10.3390/biom10081150. Biomolecules. 2020. PMID: 32781654 Free PMC article. Review. - Secretome analysis of patient-derived GBM tumor spheres identifies midkine as a potent therapeutic target.
Han S, Shin H, Lee JK, Liu Z, Rabadan R, Lee J, Shin J, Lee C, Yang H, Kim D, Kim SH, Kim J, Oh JW, Kong DS, Lee JI, Seol HJ, Choi JW, Kang HJ, Nam DH. Han S, et al. Exp Mol Med. 2019 Dec 6;51(12):1-11. doi: 10.1038/s12276-019-0351-y. Exp Mol Med. 2019. PMID: 31811117 Free PMC article. - Sustained proliferation in cancer: Mechanisms and novel therapeutic targets.
Feitelson MA, Arzumanyan A, Kulathinal RJ, Blain SW, Holcombe RF, Mahajna J, Marino M, Martinez-Chantar ML, Nawroth R, Sanchez-Garcia I, Sharma D, Saxena NK, Singh N, Vlachostergios PJ, Guo S, Honoki K, Fujii H, Georgakilas AG, Bilsland A, Amedei A, Niccolai E, Amin A, Ashraf SS, Boosani CS, Guha G, Ciriolo MR, Aquilano K, Chen S, Mohammed SI, Azmi AS, Bhakta D, Halicka D, Keith WN, Nowsheen S. Feitelson MA, et al. Semin Cancer Biol. 2015 Dec;35 Suppl(Suppl):S25-S54. doi: 10.1016/j.semcancer.2015.02.006. Epub 2015 Apr 17. Semin Cancer Biol. 2015. PMID: 25892662 Free PMC article. Review. - The Role of Estrogen and Estrogen Receptors in Head and Neck Tumors.
Kranjčević JK, Čonkaš J, Ozretić P. Kranjčević JK, et al. Cancers (Basel). 2024 Apr 19;16(8):1575. doi: 10.3390/cancers16081575. Cancers (Basel). 2024. PMID: 38672656 Free PMC article. Review. - Natural Compounds That Target DNA Repair Pathways and Their Therapeutic Potential to Counteract Cancer Cells.
Lagunas-Rangel FA, Bermúdez-Cruz RM. Lagunas-Rangel FA, et al. Front Oncol. 2020 Nov 19;10:598174. doi: 10.3389/fonc.2020.598174. eCollection 2020. Front Oncol. 2020. PMID: 33330091 Free PMC article. Review.
References
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. - PubMed
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. - PubMed
- Grady WM. Transforming growth factor-beta, Smads, and cancer. Clin Cancer Res. 2005;11:3151–54. - PubMed
- Chiao PJ, Hunt KK, Grau AM, Abramian A, Fleming J, Zhang W, et al. Tumor suppressor gene Smad4/DPC4, its downstream target genes, and regulation of cell cycle. Ann N Y Acad Sci. 1999;880:31–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UL1 RR025780/RR/NCRR NIH HHS/United States
- R01 AT003623-05/AT/NCCIH NIH HHS/United States
- UL1 RR025780-04/RR/NCRR NIH HHS/United States
- P20-CA103680/CA/NCI NIH HHS/United States
- P20 CA103680/CA/NCI NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- R01 AT003623/AT/NCCIH NIH HHS/United States
- P20 CA103680-05/CA/NCI NIH HHS/United States
- P30-CA046934/CA/NCI NIH HHS/United States
- R01 AT-003623/AT/NCCIH NIH HHS/United States
- UL1-RR-025780/RR/NCRR NIH HHS/United States
- P30 CA046934-23/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous