Independent origins of cultivated coconut (Cocos nucifera L.) in the old world tropics - PubMed (original) (raw)
Independent origins of cultivated coconut (Cocos nucifera L.) in the old world tropics
Bee F Gunn et al. PLoS One. 2011.
Abstract
As a portable source of food, water, fuel, and construction materials, the coconut (Cocos nucifera L.) played a fundamental role in human migrations and the development of civilization across the humid tropics. Here we investigated the coconut's domestication history and its population genetic structure as it relates to human dispersal patterns. A sample of 1,322 coconut accessions, representing the geographical and phenotypic diversity of the species, was examined using ten microsatellite loci. Bayesian analyses reveal two highly genetically differentiated subpopulations that correspond to the Pacific and Indo-Atlantic oceanic basins. This pattern suggests independent origins of coconut cultivation in these two world regions, with persistent population structure on a global scale despite long-term human cultivation and dispersal. Pacific coconuts show additional genetic substructure corresponding to phenotypic and geographical subgroups; moreover, the traits that are most clearly associated with selection under human cultivation (dwarf habit, self-pollination, and "niu vai" fruit morphology) arose only in the Pacific. Coconuts that show evidence of genetic admixture between the Pacific and Indo-Atlantic groups occur primarily in the southwestern Indian Ocean. This pattern is consistent with human introductions of Pacific coconuts along the ancient Austronesian trade route connecting Madagascar to Southeast Asia. Admixture in coastal east Africa may also reflect later historic Arab trading along the Indian Ocean coastline. We propose two geographical origins of coconut cultivation: island Southeast Asia and southern margins of the Indian subcontinent.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Results of Structure analysis for a worldwide sample of 1322 coconuts.
Population assignments for each accession are shown at K = 2 subpopulations. Numbers along the x-axis correspond to group designations in Table 1. Vertical black lines distinguish the population groups.
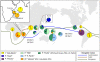
Figure 2. Geographical distributions of Indo-Atlantic and Pacific coconut subpopulations.
Subpopulation designations correspond to assignments at Q≥80% membership in Structure analyses at K = 5. ‘I’ and ‘P’ prefixes in the legend indicate ‘Indo-Atlantic’ and ‘Pacific’ population assignments at K = 2 assumed populations (≥80% membership; see Fig. 1). Lines indicate proposed coconut dispersal routes by humans. Pie chart labels correspond to the following countries (ISO abbreviations) and sample sizes: A = IND, LKA, SEY (114); B = BEN, CIV, CMR, GHA (29); C = JAM, MEX (Atlantic) (13); D = BRA (72); E = KEN, MOZ, TZA (116); F = MAD, COM (65); G = Dwarf (54); H = CHN, KHM, MYS, THD, VNM (66); I = IDN (25); J = PHL (46); K = PAN (105); L = MEX (Pacific) (43); M = PNG (141); N = KIT, MHL, TUV (43); O = NCL, SLB, VUT (360); P = COK, FJI, PYF (30). Inset: subpopulation compositions for Madagascar, Comoros, and Seychelles. Pie chart composition is selected to reflect geographical population structure and does not correspond directly to GPC/CIRAD designations in Table 1.
Similar articles
- QTL analysis of fruit components in the progeny of a Rennell Island Tall coconut (Cocos nucifera L.) individual.
Baudouin L, Lebrun P, Konan JL, Ritter E, Berger A, Billotte N. Baudouin L, et al. Theor Appl Genet. 2006 Jan;112(2):258-68. doi: 10.1007/s00122-005-0123-z. Epub 2005 Nov 24. Theor Appl Genet. 2006. PMID: 16307230 - The genome draft of coconut (Cocos nucifera).
Xiao Y, Xu P, Fan H, Baudouin L, Xia W, Bocs S, Xu J, Li Q, Guo A, Zhou L, Li J, Wu Y, Ma Z, Armero A, Issali AE, Liu N, Peng M, Yang Y. Xiao Y, et al. Gigascience. 2017 Nov 1;6(11):1-11. doi: 10.1093/gigascience/gix095. Gigascience. 2017. PMID: 29048487 Free PMC article. - Kopyor versus macapuno coconuts: are these two edible mutants of Southeast Asia the same?
Wicaksono A, Raihandhany R, Teixeira da Silva JA. Wicaksono A, et al. Planta. 2021 Sep 28;254(5):86. doi: 10.1007/s00425-021-03740-y. Planta. 2021. PMID: 34585305 Review. - The human genetic history of South Asia.
Majumder PP. Majumder PP. Curr Biol. 2010 Feb 23;20(4):R184-7. doi: 10.1016/j.cub.2009.11.053. Curr Biol. 2010. PMID: 20178765 Review.
Cited by
- The presence of coconut in southern Panama in pre-Columbian times: clearing up the confusion.
Baudouin L, Gunn BF, Olsen KM. Baudouin L, et al. Ann Bot. 2014 Jan;113(1):1-5. doi: 10.1093/aob/mct244. Epub 2013 Nov 13. Ann Bot. 2014. PMID: 24227445 Free PMC article. - Genetic Relationships among Tall Coconut Palm (Cocos nucifera L.) Accessions of the International Coconut Genebank for Latin America and the Caribbean (ICG-LAC), Evaluated Using Microsatellite Markers (SSRs).
Loiola CM, Azevedo AO, Diniz LE, Aragão WM, Azevedo CD, Santos PH, Ramos HC, Pereira MG, Ramos SR. Loiola CM, et al. PLoS One. 2016 Mar 14;11(3):e0151309. doi: 10.1371/journal.pone.0151309. eCollection 2016. PLoS One. 2016. PMID: 26974540 Free PMC article. - Mining and validation of novel genotyping-by-sequencing (GBS)-based simple sequence repeats (SSRs) and their application for the estimation of the genetic diversity and population structure of coconuts (Cocos nucifera L.) in Thailand.
Riangwong K, Wanchana S, Aesomnuk W, Saensuk C, Nubankoh P, Ruanjaichon V, Kraithong T, Toojinda T, Vanavichit A, Arikit S. Riangwong K, et al. Hortic Res. 2020 Oct 1;7:156. doi: 10.1038/s41438-020-00374-1. eCollection 2020. Hortic Res. 2020. PMID: 33082963 Free PMC article. - Genetic relationship and diversity among coconut (Cocos nucifera L.) accessions revealed through SCoT analysis.
Rajesh MK, Sabana AA, Rachana KE, Rahman S, Jerard BA, Karun A. Rajesh MK, et al. 3 Biotech. 2015 Dec;5(6):999-1006. doi: 10.1007/s13205-015-0304-7. Epub 2015 May 7. 3 Biotech. 2015. PMID: 28324407 Free PMC article. - Complete sequence and comparative analysis of the chloroplast genome of coconut palm (Cocos nucifera).
Huang YY, Matzke AJ, Matzke M. Huang YY, et al. PLoS One. 2013 Aug 30;8(8):e74736. doi: 10.1371/journal.pone.0074736. eCollection 2013. PLoS One. 2013. PMID: 24023703 Free PMC article.
References
- Harries HC. The evolution, dissemination and classification of Cocos nucifera. L. Bot Rev. 1978;44:265–319.
- Zizumbo-Villarreal D. History of coconut (Cocos nucifera L.) in Mexico: 1539–1810. Genet Resour Crop Evol. 1996;43:505–515.
- Batugal P, Rao V, Oliver J, editors. Coconut Genetic Resources. 2005. IPGRI - Regional Office for Asia, the Pacific and Oceania, Serdang, Malaysia.
- Ward G, Brookfield M. The dispersal of the coconut: did it float or was it carried to Panama? J Biogeogr. 1992;19:467–480.
- Lebrun P, N'Cho Y, Bourdeix R, Baudouin L. Coconut. In: Hamon P, Seguin M, Perrier X, Glaszmann JC, editors. Genetic diversity of cultivated tropical plants. Paris, France: Science Publishers, Inc. and CIRAD, France; 2003. pp. 219–238.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials