Inference of human population history from individual whole-genome sequences - PubMed (original) (raw)
Inference of human population history from individual whole-genome sequences
Heng Li et al. Nature. 2011.
Abstract
The history of human population size is important for understanding human evolution. Various studies have found evidence for a founder event (bottleneck) in East Asian and European populations, associated with the human dispersal out-of-Africa event around 60 thousand years (kyr) ago. However, these studies have had to assume simplified demographic models with few parameters, and they do not provide a precise date for the start and stop times of the bottleneck. Here, with fewer assumptions on population size changes, we present a more detailed history of human population sizes between approximately ten thousand and a million years ago, using the pairwise sequentially Markovian coalescent model applied to the complete diploid genome sequences of a Chinese male (YH), a Korean male (SJK), three European individuals (J. C. Venter, NA12891 and NA12878 (ref. 9)) and two Yoruba males (NA18507 (ref. 10) and NA19239). We infer that European and Chinese populations had very similar population-size histories before 10-20 kyr ago. Both populations experienced a severe bottleneck 10-60 kyr ago, whereas African populations experienced a milder bottleneck from which they recovered earlier. All three populations have an elevated effective population size between 60 and 250 kyr ago, possibly due to population substructure. We also infer that the differentiation of genetically modern humans may have started as early as 100-120 kyr ago, but considerable genetic exchanges may still have occurred until 20-40 kyr ago.
Figures
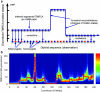
Figure 1. Illustration of the PSMC model and its application to simulated data
(a) The PSMC infers the local time to the most recent common ancestor (TMRCA) based on the local density of heterozygotes, using a Hidden Markov Model, where the observation is a diploid sequence, the hidden states are discretized TMRCA and the transitions represent ancestral recombination events. (b) We used the ms software to simulate the TMRCA relating the two alleles of an individual across a 200kb region (the thick red line), and inferred the local TMRCA at each locus using the PSMC (the heat map). The inference usually includes the correct time, with the greatest errors at transition points.
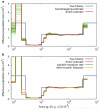
Figure 2. PSMC estimate on simulated data
(a) PSMC estimate on data simulated by msHOT. The blue curve is the population size history used in simulation; the red curve is the PSMC estimate on the originally simulated sequence; the 100 thin green curves are the PSMC estimates on 100 sequences randomly resampled from the original sequence. (b) PSMC estimate on data with variable mutation rate or with hotspots.
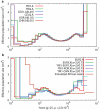
Figure 3. PSMC estimate on real data
(a) The population sizes inferred from autosomes of six individuals. 5%, 10% and 29% of heterozygotes are assumed to be missing in CHN.A, KOR.A and EUR1.A, respectively. (b) The population sizes inferred from male-combined X chromosomes and the simulated African-Asian combined sequences from the best-fit model by Schaffner et al. Sizes inferred from X chromosome data are scaled by 4/3; the neutral mutation rate on X, which is used in time scaling, is estimated with the ratio of male-to-female mutation rate α equal to 2 (Methods).
Similar articles
- PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers.
Nadachowska-Brzyska K, Burri R, Smeds L, Ellegren H. Nadachowska-Brzyska K, et al. Mol Ecol. 2016 Mar;25(5):1058-72. doi: 10.1111/mec.13540. Epub 2016 Feb 15. Mol Ecol. 2016. PMID: 26797914 Free PMC article. - Demographic inference in barn swallows using whole-genome data shows signal for bottleneck and subspecies differentiation during the Holocene.
Smith CCR, Flaxman SM, Scordato ESC, Kane NC, Hund AK, Sheta BM, Safran RJ. Smith CCR, et al. Mol Ecol. 2018 Nov;27(21):4200-4212. doi: 10.1111/mec.14854. Epub 2018 Sep 22. Mol Ecol. 2018. PMID: 30176075 - Estimating variable effective population sizes from multiple genomes: a sequentially markov conditional sampling distribution approach.
Sheehan S, Harris K, Song YS. Sheehan S, et al. Genetics. 2013 Jul;194(3):647-62. doi: 10.1534/genetics.112.149096. Epub 2013 Apr 22. Genetics. 2013. PMID: 23608192 Free PMC article. - Ancestral Population Genomics.
Dutheil JY, Hobolth A. Dutheil JY, et al. Methods Mol Biol. 2019;1910:555-589. doi: 10.1007/978-1-4939-9074-0_18. Methods Mol Biol. 2019. PMID: 31278677 Review. - [The origin and evolution history of East Asian populations from genetic perspectives].
Tian JY, Li YC, Kong QP, Zhang YP. Tian JY, et al. Yi Chuan. 2018 Oct 20;40(10):814-824. doi: 10.16288/j.yczz.18-202. Yi Chuan. 2018. PMID: 30369466 Review. Chinese.
Cited by
- Revealing the Demographic History of the European Nightjar (Caprimulgus europaeus).
Day G, Fox G, Hipperson H, Maher KH, Tucker R, Horsburgh GJ, Waters D, Durant KL, Burke T, Slate J, Arnold KE. Day G, et al. Ecol Evol. 2024 Oct 26;14(10):e70460. doi: 10.1002/ece3.70460. eCollection 2024 Oct. Ecol Evol. 2024. PMID: 39463738 Free PMC article. - Infanticide and Human Self Domestication.
Kimbrough EO, Myers GM, Robson AJ. Kimbrough EO, et al. Front Psychol. 2021 May 5;12:667334. doi: 10.3389/fpsyg.2021.667334. eCollection 2021. Front Psychol. 2021. PMID: 34025530 Free PMC article. No abstract available. - Insights into the demographic history of Asia from common ancestry and admixture in the genomic landscape of present-day Austroasiatic speakers.
Tagore D, Aghakhanian F, Naidu R, Phipps ME, Basu A. Tagore D, et al. BMC Biol. 2021 Mar 29;19(1):61. doi: 10.1186/s12915-021-00981-x. BMC Biol. 2021. PMID: 33781248 Free PMC article. - The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population.
Lack JB, Cardeno CM, Crepeau MW, Taylor W, Corbett-Detig RB, Stevens KA, Langley CH, Pool JE. Lack JB, et al. Genetics. 2015 Apr;199(4):1229-41. doi: 10.1534/genetics.115.174664. Epub 2015 Jan 27. Genetics. 2015. PMID: 25631317 Free PMC article. - A world in a grain of sand: human history from genetic data.
Colonna V, Pagani L, Xue Y, Tyler-Smith C. Colonna V, et al. Genome Biol. 2011 Nov 21;12(11):234. doi: 10.1186/gb-2011-12-11-234. Genome Biol. 2011. PMID: 22104725 Free PMC article. Review.
References
References for online methods
- Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb Symp Quant Biol. 1987;52:863–867. - PubMed
References
- Reich DE, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases