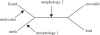MicroRNAs support a turtle + lizard clade - PubMed (original) (raw)
Comparative Study
. 2012 Feb 23;8(1):104-7.
doi: 10.1098/rsbl.2011.0477. Epub 2011 Jul 20.
Affiliations
- PMID: 21775315
- PMCID: PMC3259949
- DOI: 10.1098/rsbl.2011.0477
Comparative Study
MicroRNAs support a turtle + lizard clade
Tyler R Lyson et al. Biol Lett. 2012.
Abstract
Despite much interest in amniote systematics, the origin of turtles remains elusive. Traditional morphological phylogenetic analyses place turtles outside Diapsida-amniotes whose ancestor had two fenestrae in the temporal region of the skull (among the living forms the tuatara, lizards, birds and crocodilians)-and allied with some unfenestrate-skulled (anapsid) taxa. Nonetheless, some morphological analyses place turtles within Diapsida, allied with Lepidosauria (tuatara and lizards). Most molecular studies agree that turtles are diapsids, but rather than allying them with lepidosaurs, instead place turtles near or within Archosauria (crocodilians and birds). Thus, three basic phylogenetic positions for turtles with respect to extant Diapsida are currently debated: (i) sister to Diapsida, (ii) sister to Lepidosauria, or (iii) sister to, or within, Archosauria. Interestingly, although these three alternatives are consistent with a single unrooted four-taxon tree for extant reptiles, they differ with respect to the position of the root. Here, we apply a novel molecular dataset, the presence versus absence of specific microRNAs, to the problem of the phylogenetic position of turtles and the root of the reptilian tree, and find that this dataset unambiguously supports a turtle + lepidosaur group. We find that turtles and lizards share four unique miRNA gene families that are not found in any other organisms' genome or small RNA library, and no miRNAs are found in all diapsids but not turtles, or in turtles and archosaurs but not in lizards. The concordance between our result and some morphological analyses suggests that there have been numerous morphological convergences and reversals in reptile phylogeny, including the loss of temporal fenestrae.
Figures
Figure 1.
The inter-relationships among the major groups of reptiles. Using an unrooted tree, it is possible to show that although each of the three previous hypotheses concerning turtle relationships—morphology 1 [–3], morphology 2 [–7] and the molecular results [–12]—agree on the topology, they disagree on the position of the root (arrows).
Figure 2.
microRNAs support a turtle–lizard relationship. (a) Structures and alignments of the mature sequences for three of the 35 families analysed phylogenetically, the reptile-specific miR-1677, the archosaur-specific miR-1791 and miR-5390, a novel miRNA shared between Anolis carolinensis and C. pictus. Mature sequences within the context of the miRNA hairpin are shown in grey; changes in the mature sequence with respect to the reference sequence, either chicken or lizard, are shown in bold. (b) To arbitrate among these competing hypotheses (see figure 1), eight amniote taxa were scored for the presence/absence of 35 miRNA families with the frog Xenopus laevis as the outgroup. Using a combination of small RNA library sequencing coupled with genomic searches (electronic supplementary material, file S1), we find that the turtle C. picta shares four miRNA families with the lizard A. carolinensis that are not found elsewhere in the animal kingdom, supporting the rooting position of ‘morphology 2’ in figure 1 [–7]. Bremer support indexes are indicated at each node. Character changes on branch: grey boxes, acquisition of miRNA family; filled triangle, loss of miRNA family.
Similar articles
- Toward consilience in reptile phylogeny: miRNAs support an archosaur, not lepidosaur, affinity for turtles.
Field DJ, Gauthier JA, King BL, Pisani D, Lyson TR, Peterson KJ. Field DJ, et al. Evol Dev. 2014 Jul-Aug;16(4):189-96. doi: 10.1111/ede.12081. Epub 2014 May 5. Evol Dev. 2014. PMID: 24798503 Free PMC article. - Complete mitochondrial DNA sequences of the green turtle and blue-tailed mole skink: statistical evidence for archosaurian affinity of turtles.
Kumazawa Y, Nishida M. Kumazawa Y, et al. Mol Biol Evol. 1999 Jun;16(6):784-92. doi: 10.1093/oxfordjournals.molbev.a026163. Mol Biol Evol. 1999. PMID: 10368956 - Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria).
Chiari Y, Cahais V, Galtier N, Delsuc F. Chiari Y, et al. BMC Biol. 2012 Jul 27;10:65. doi: 10.1186/1741-7007-10-65. BMC Biol. 2012. PMID: 22839781 Free PMC article. - The evolutionary position of turtles revised.
Zardoya R, Meyer A. Zardoya R, et al. Naturwissenschaften. 2001 May;88(5):193-200. doi: 10.1007/s001140100228. Naturwissenschaften. 2001. PMID: 11482432 Review.
Cited by
- Building a robust a-p axis.
Heimberg A, McGlinn E. Heimberg A, et al. Curr Genomics. 2012 Jun;13(4):278-88. doi: 10.2174/138920212800793348. Curr Genomics. 2012. PMID: 23204917 Free PMC article. - Using genes as characters and a parsimony analysis to explore the phylogenetic position of turtles.
Lu B, Yang W, Dai Q, Fu J. Lu B, et al. PLoS One. 2013 Nov 21;8(11):e79348. doi: 10.1371/journal.pone.0079348. eCollection 2013. PLoS One. 2013. PMID: 24278129 Free PMC article. - Characterization of Two Transposable Elements and an Ultra-Conserved Element Isolated in the Genome of Zootoca vivipara (Squamata, Lacertidae).
Mezzasalma M, Capriglione T, Kupriyanova L, Odierna G, Pallotta MM, Petraccioli A, Picariello O, Guarino FM. Mezzasalma M, et al. Life (Basel). 2023 Feb 24;13(3):637. doi: 10.3390/life13030637. Life (Basel). 2023. PMID: 36983793 Free PMC article. - A comparative analysis of heart microRNAs in vertebrates brings novel insights into the evolution of genetic regulatory networks.
Nachtigall PG, Bovolenta LA, Patton JG, Fromm B, Lemke N, Pinhal D. Nachtigall PG, et al. BMC Genomics. 2021 Mar 4;22(1):153. doi: 10.1186/s12864-021-07441-4. BMC Genomics. 2021. PMID: 33663371 Free PMC article. - A critical appraisal of the use of microRNA data in phylogenetics.
Thomson RC, Plachetzki DC, Mahler DL, Moore BR. Thomson RC, et al. Proc Natl Acad Sci U S A. 2014 Sep 2;111(35):E3659-68. doi: 10.1073/pnas.1407207111. Epub 2014 Jul 28. Proc Natl Acad Sci U S A. 2014. PMID: 25071211 Free PMC article.
References
- Gauthier J., Kluge A. G., Rowe T. 1988. Amniote phylogeny and the importance of fossils. Cladistics 4, 105–20910.1111/j.1096-0031.1988.tb00514.x (doi:10.1111/j.1096-0031.1988.tb00514.x) - DOI - DOI - PubMed
- Lee M. S. Y. 1993. The origin of the turtle body plan: bridging a famous morphological gap. Science 261, 1716–172010.1126/science.261.5129.1716 (doi:10.1126/science.261.5129.1716) - DOI - DOI - PubMed
- Lyson T. R., Bever G. S., Bhullar B.-A. S., Joyce W. G., Gauthier J. A. 2010. Transitional fossils and the origin of turtles. Biol. Lett. 6, 830–83310.1098/rsbl.2010.0371 (doi:10.1098/rsbl.2010.0371) - DOI - DOI - PMC - PubMed
- Rieppel O., deBraga M. 1996. Turtles as diapsid reptiles. Nature 384, 453–45510.1038/384453a0 (doi:10.1038/384453a0) - DOI - DOI
- Müller J. 2004. The relationships among diapsid reptiles and the influence of taxon selection. In Recent advances in the origin and early radiation of vertebrates (eds Arratia M. V. H., Cloutier R.), pp. 379–408 München, Germany: Verlag Dr. Friedrich Pfeil
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources