Expression profiling of the intermediate and late stages of poxvirus replication - PubMed (original) (raw)
Expression profiling of the intermediate and late stages of poxvirus replication
Zhilong Yang et al. J Virol. 2011 Oct.
Abstract
The double-stranded DNA genome of vaccinia virus (VACV), the prototype poxvirus, contains approximately 200 open reading frames (ORFs) that are transcribed at early, intermediate, and late stages of infection. Previous high-throughput deep RNA sequencing allowed us to map 118 VACV early genes that are expressed before viral DNA replication and 93 postreplicative genes. However, the intermediate- and late-stage postreplicative genes could not be differentiated. Here, we synchronized infections with a reversible inhibitor of DNA replication and used a VACV mutant that conditionally transcribes late genes to sequence the two classes of mRNAs. In addition, each postreplicative ORF was individually expressed under conditions that distinguished intermediate and late classes. We identified 38 VACV genes that belong to the late class and 53 that belong to the intermediate class, with some of the latter continuing to be expressed late. These data allowed us to prepare a genome-wide early, intermediate, and late transcription map. Inspection of sequences upstream of these ORFs revealed distinctive characteristics of intermediate and late promoters and suggested that some promoters have intermediate and late elements. The intermediate genes encoded many DNA binding/packaging and core-associated proteins in addition to late transcription factors; the late genes encoded many morphogenesis and mature virion membrane proteins, including those involved in entry, in addition to early transcription factors. The top-ranked antigens for CD4(+) T cells and B cells were mainly intermediate rather than late gene products. The differentiation of intermediate and late genes may enhance understanding of poxvirus replication and lead to improvements in expression vectors and recombinant vaccines.
Figures
Fig. 1.
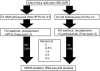
Flowchart for intermediate (Int) and late mRNA analyses. HeLa S3 cells infected with vRO-G8R at a multiplicity of infection of 20 PFU per cell were incubated for 4 h in the presence of 10 mM HU to inhibit DNA replication with or without 5 mM IPTG. Synchronization of the infection was achieved by washing the cells to remove HU and resuspending them in the presence of CHX to prevent late gene expression or in the presence of IPTG to allow late gene expression. The cells were harvested at 0, 0.5, 1, 2, and 4 h for RNA isolation and sequencing.
Fig. 2.
Temporal expression of VACV mRNAs following removal of HU. HeLa cells were infected with vRO-G8R in the absence or presence of IPTG as outlined in Fig. 1. After removal of HU, polyadenylated RNA was isolated at 0, 0.5, 1, 2, and 4 h, and cDNAs were made and sequenced. Read counts per nucleotide were plotted along the complete VACV ORF map. Counts above and below the line represent RNAs transcribed from the top DNA strand in the rightward direction and from the bottom strand in the leftward direction, respectively. Only the 1-, 2-, and 4-h data are shown here; the 0- and 0.5-h data are shown in Fig. S1 in the supplemental material. ORFs are indicated by arrows; the letters at the bottom represent HindIII fragments. (A) Cells without IPTG and with CHX. (B) Cells with IPTG and without CHX.
Fig. 3.
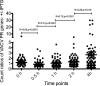
Ratios (+IPTG/−IPTG) of read counts mapping to VACV postreplicative ORFs. For each time point, the reads mapping to early mRNAs were removed, and the reads mapping to postreplicative ORFs were normalized by the total read counts. The ratios (+IPTG/−IPTG) of read counts were plotted. The horizontal lines are the mean ratios with the standard error of the mean at each time. R is the correlation coefficient between two neighboring sets of ratios; a smaller R signifies a change in ratios and correlates with the presence of late mRNAs in the sample with IPTG at 4 h. The P values are the chance that random ratios would result in a correlation coefficient as far from zero (or further) as observed if there really is no correlation.
Fig. 4.
Superimposed VACV genome-wide transcriptome maps at 4 h after HU removal. The data obtained at 4 h after HU removal, as described in the legend of Fig. 2, were plotted as the number of reads per nucleotide and expanded. The color code is as follows: red, reads with IPTG; green, reads without IPTG; yellow, superimposed reads.
Fig. 5.
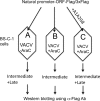
Scheme for distinguishing intermediate and late genes by transfection. BS-C-1 cells were infected with VACV in the absence (A) or presence (B and C) of AraC and transfected with a single plasmid containing a Flag-tagged ORF under its natural intermediate or late promoter (A and B) or cotransfected with the latter plasmid and plasmid A1A2G8 (C) containing the three late transcription factors under their natural intermediate promoters. The lysates were analyzed by Western blotting with antibody to the Flag epitope (α-Flag Ab). In the absence of AraC, both intermediate and late genes can be expressed; in the presence of AraC only intermediate genes can be expressed since late transcription factors are not made. However, late gene expression in the presence of AraC can be rescued by cotransfection of a plasmid expressing the three late transcription factors. In a related assay (not shown here) the VACV ORF was replaced by EGFP under the control of the putative intermediate or late promoter.
Fig. 6.
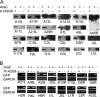
Expression of individual genes in the presence and absence of AraC. Employing the scheme shown in Fig. 5, each Flag- or 3×Flag-tagged VACV postreplicative gene in a plasmid was transfected into VACV-infected BS-C-1 cells in the presence (+) or absence (−) of AraC. Western blots were probed with anti-Flag M2 antibody. In most cases, a single band of the expected size was discerned, and only a small part of the Western blot is shown. An expanded segment of the Western blot is shown in cases where more than one band is present due to protein processing, glycosylation, or SDS-resistant aggregates. Where applicable, the common (Copenhagen) HindIII fragment letter/number was used as the ORF name; otherwise the WR number was used. The protein expression patterns are as follows: I, robust expression only in the absence of AraC; IIA, robust expression in the absence and presence of AraC; IIB, higher expression in the presence of AraC than in the absence.
Fig. 7.
Rescue of late gene expression by cotransfection of late transcription factors. As described in the scheme of Fig. 5, late gene expression was rescued by expressing late transcription factors in the presence of AraC. (A) BS-C-1 cells were infected with VACV in the presence (+) or absence (−) of AraC and in each case transfected with a plasmid containing a Flag- or 3×Flag-tagged VACV postreplicative gene with its natural promoter. The cells were cotransfected with A1A2G8 (+) or the vector plasmid (−). The cells were lysed, and Western blots were probed with anti-Flag M2 antibody. (B) The protocol was similar to that of panel A except that the plasmid encoded EGFP under the VACV promoter instead of the Flag-tagged ORF. The Western blot was probed with anti-EGFP antibody and with anti-GAPDH antibody as a loading control. In both panels, the common (Copenhagen) HindIII fragment letter/number was used as the ORF name.
Fig. 8.
VACV transcriptome map. VACV WR ORFs are shown as colored arrows indicating the direction of transcription. When applicable, the common (Copenhagen) HindIII fragment letter/number name was used to identify ORFs; otherwise the VACV WR name was provided. The numbers from 1 to 194711 indicate the nucleotide positions on the VACV genome. Note that each ORF has been assigned the stage at which its earliest expression can be detected and that the presence of additional promoter elements that potentially contribute to later stages of gene expression are not indicated.
Fig. 9.
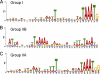
Motif logos of VACV promoters. Motifs were generated by the program MEME using sequences spanning 50 nt upstream and 4 nt downstream of the start codons. The search parameters were one motif per sequence and a motif length of 33 to 38 nt. Motifs represent late genes (A), intermediate genes (B), and intermediate genes with late expression (C), as shown in Fig. 6.
Fig. 10.
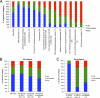
Gene functions and antigenicity of different expression classes. (A) The individual genes in each functional category are listed in Table S1 in the supplemental material. The numbers of genes in each category are indicated. (B) CD8+, CD4+, and B cell antigens were obtained from a recent review (19) and were grouped by expression class according to Table S1. The numbers following each category indicate the number of genes in that category. (C) Highly recognized CD8+ T cell (G5.5R, G5R, E2L, A19L, D12L, F12L, C7L, A47L, A55R, and A48R), CD4+ T cell (D13L, H3L, L4R, A2.5L, B2R, A9L, A26L, D8L, F13L, H7R, I1L, J6R, and O3L), and B cell (A10L, H3L, B5R, A33R, A27L, A56R, A25L, D13L D8L, and A13L) antigens, from the same review as in panel B, were grouped by expression class.
Similar articles
- Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing.
Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. Yang Z, et al. Proc Natl Acad Sci U S A. 2010 Jun 22;107(25):11513-8. doi: 10.1073/pnas.1006594107. Epub 2010 Jun 7. Proc Natl Acad Sci U S A. 2010. PMID: 20534518 Free PMC article. - Novel Nonreplicating Vaccinia Virus Vector Enhances Expression of Heterologous Genes and Suppresses Synthesis of Endogenous Viral Proteins.
Wyatt LS, Xiao W, Americo JL, Earl PL, Moss B. Wyatt LS, et al. mBio. 2017 Jun 6;8(3):e00790-17. doi: 10.1128/mBio.00790-17. mBio. 2017. PMID: 28588133 Free PMC article. - Drosophila S2 cells are non-permissive for vaccinia virus DNA replication following entry via low pH-dependent endocytosis and early transcription.
Bengali Z, Satheshkumar PS, Yang Z, Weisberg AS, Paran N, Moss B. Bengali Z, et al. PLoS One. 2011 Feb 15;6(2):e17248. doi: 10.1371/journal.pone.0017248. PLoS One. 2011. PMID: 21347205 Free PMC article. - Organization and expression of the poxvirus genome.
Wittek R. Wittek R. Experientia. 1982 Mar 15;38(3):285-97. doi: 10.1007/BF01949349. Experientia. 1982. PMID: 6281055 Review. - Transcription in vaccinia virus.
Kent R. Kent R. Microbiologia. 1989 Sep;5(2):69-77. Microbiologia. 1989. PMID: 2698178 Review.
Cited by
- Engineering Oncolytic Vaccinia Virus to redirect Macrophages to Tumor Cells.
Cao F, Nguyen P, Hong B, DeRenzo C, Rainusso NC, Rodriguez Cruz T, Wu MF, Liu H, Song XT, Suzuki M, Wang LL, Yustein JT, Gottschalk S. Cao F, et al. Adv Cell Gene Ther. 2021 Apr;4(2):e99. doi: 10.1002/acg2.99. Epub 2020 Jul 3. Adv Cell Gene Ther. 2021. PMID: 33829146 Free PMC article. - RACK1 Regulates Poxvirus Protein Synthesis Independently of Its Role in Ribosome-Based Stress Signaling.
Park C, Walsh D. Park C, et al. J Virol. 2022 Sep 28;96(18):e0109322. doi: 10.1128/jvi.01093-22. Epub 2022 Sep 13. J Virol. 2022. PMID: 36098514 Free PMC article. - Multi-omics characterization of the monkeypox virus infection.
Huang Y, Bergant V, Grass V, Emslander Q, Hamad MS, Hubel P, Mergner J, Piras A, Krey K, Henrici A, Öllinger R, Tesfamariam YM, Dalla Rosa I, Bunse T, Sutter G, Ebert G, Schmidt FI, Way M, Rad R, Bowie AG, Protzer U, Pichlmair A. Huang Y, et al. Nat Commun. 2024 Aug 8;15(1):6778. doi: 10.1038/s41467-024-51074-6. Nat Commun. 2024. PMID: 39117661 Free PMC article. - Reverse genetics analysis of poxvirus intermediate transcription factors.
Warren RD, Cotter CA, Moss B. Warren RD, et al. J Virol. 2012 Sep;86(17):9514-9. doi: 10.1128/JVI.06902-11. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740406 Free PMC article. - Long-read assays shed new light on the transcriptome complexity of a viral pathogen.
Tombácz D, Prazsák I, Csabai Z, Moldován N, Dénes B, Snyder M, Boldogkői Z. Tombácz D, et al. Sci Rep. 2020 Aug 14;10(1):13822. doi: 10.1038/s41598-020-70794-5. Sci Rep. 2020. PMID: 32796917 Free PMC article.
References
- Bailey T. L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials