Creating a "hopeful monster": mouse forward genetic screens - PubMed (original) (raw)
Creating a "hopeful monster": mouse forward genetic screens
Vanessa L Horner et al. Methods Mol Biol. 2011.
Abstract
One of the most straightforward approaches to making novel biological discoveries is the forward genetic screen. The time is ripe for forward genetic screens in the mouse since the mouse genome is sequenced, but the function of many of the genes remains unknown. Today, with careful planning, such screens are within the reach of even small individual labs. In this chapter we first discuss the types of screens in existence, as well as how to design a screen to recover mutations that are relevant to the interests of a lab. We then describe how to create mutations using the chemical N-ethyl-N-nitrosourea (ENU), including a detailed injection protocol. Next, we outline breeding schemes to establish mutant lines for each type of screen. Finally, we explain how to map mutations using recombination and how to ensure that a particular mutation causes a phenotype. Our goal is to make forward genetics in the mouse accessible to any lab with the desire to do it.
Figures
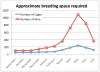
Figure 11.1
Approximate breeding space required per month in a generic yearlong screen for recessive mutations.
Figure 11.2
Classes of genome-wide and region-specific forward genetic screens. Adapted by permission from Macmillan Publishers Ltd: Nat Rev Genet (33), copyright 2005.
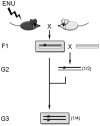
Figure 11.3
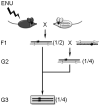
Crossing scheme for dominant (upper grey box) or recessive (lower grey box) mutant alleles. In this and all subsequent figures, chromosomes from the mutagenized black mouse are represented as black bars; chromosomes from the white mouse are represented as white bars. Additionally, in all figures stars represent mutant alleles.
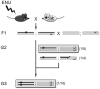
Figure 11.4
Crossing scheme for dominant (upper grey box) or recessive (lower grey box) modifier alleles. In this crossing scheme the allele to be modified (in the white mouse) is assumed to be homozygous lethal or sterile. The half-black/half-white chromosome in the second generation indicates that either allele is acceptable in this cross.
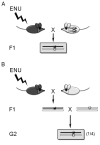
Figure 11.5
Non-complementation crossing scheme. (A) The allele to be tested (in white mouse) is viable and fertile as a homozygote. (B) The allele to be tested (in white mouse) is lethal or sterile as a homozygote.
Figure 11.6
Deletion screen crossing scheme. The chromosome with a gap indicates the region that is deleted. Mutant alleles are recovered in the second generation (grey box).
Figure 11.7
Balancer screen crossing scheme. The white bar with double-sided arrows indicates the balanced chromosome. In the first generation, F1 mice are crossed to mice carrying the inversion in trans to a WT chromosome marked with a dominant visible mutation (dotted bar with black circle). Mutant alleles are recovered in the third generation (grey box).
Similar articles
- Phenotype-driven mouse ENU mutagenesis screens.
Caspary T. Caspary T. Methods Enzymol. 2010;477:313-27. doi: 10.1016/S0076-6879(10)77016-6. Methods Enzymol. 2010. PMID: 20699148 - Forward Genetic Screen Using Zebrafish to Identify New Genes Involved in Myelination.
Kegel L, Rubio M, Almeida RG, Benito S, Klingseisen A, Lyons DA. Kegel L, et al. Methods Mol Biol. 2019;1936:185-209. doi: 10.1007/978-1-4939-9072-6_11. Methods Mol Biol. 2019. PMID: 30820900 - Forward genetic screens to identify circadian rhythm mutants in mice.
Siepka SM, Takahashi JS. Siepka SM, et al. Methods Enzymol. 2005;393:219-29. doi: 10.1016/S0076-6879(05)93007-3. Methods Enzymol. 2005. PMID: 15817290 Free PMC article. - N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express.
Cordes SP. Cordes SP. Microbiol Mol Biol Rev. 2005 Sep;69(3):426-39. doi: 10.1128/MMBR.69.3.426-439.2005. Microbiol Mol Biol Rev. 2005. PMID: 16148305 Free PMC article. Review. - Research Techniques Made Simple: Forward Genetic Screening to Uncover Genes Involved in Skin Biology.
McAlpine W, Russell J, Murray AR, Beutler B, Turer E. McAlpine W, et al. J Invest Dermatol. 2019 Sep;139(9):1848-1853.e1. doi: 10.1016/j.jid.2019.04.013. J Invest Dermatol. 2019. PMID: 31445571 Free PMC article. Review.
Cited by
- After GWAS: mice to the rescue?
Ermann J, Glimcher LH. Ermann J, et al. Curr Opin Immunol. 2012 Oct;24(5):564-70. doi: 10.1016/j.coi.2012.09.005. Epub 2012 Sep 29. Curr Opin Immunol. 2012. PMID: 23031443 Free PMC article. Review. - The logic of monsters: development and morphological diversity in stem-cell-based embryo models.
Cao D, Garai S, DiFrisco J, Veenvliet JV. Cao D, et al. Interface Focus. 2024 Oct 25;14(5):20240023. doi: 10.1098/rsfs.2024.0023. eCollection 2024 Oct 11. Interface Focus. 2024. PMID: 39464644 Free PMC article. Review. - ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2.
Jain D, Puno MR, Meydan C, Lailler N, Mason CE, Lima CD, Anderson KV, Keeney S. Jain D, et al. Elife. 2018 Jan 23;7:e30919. doi: 10.7554/eLife.30919. Elife. 2018. PMID: 29360036 Free PMC article. - A most formidable arsenal: genetic technologies for building a better mouse.
Clark JF, Dinsmore CJ, Soriano P. Clark JF, et al. Genes Dev. 2020 Oct 1;34(19-20):1256-1286. doi: 10.1101/gad.342089.120. Genes Dev. 2020. PMID: 33004485 Free PMC article. Review. - Detection of functional protein domains by unbiased genome-wide forward genetic screening.
Herzog M, Puddu F, Coates J, Geisler N, Forment JV, Jackson SP. Herzog M, et al. Sci Rep. 2018 Apr 18;8(1):6161. doi: 10.1038/s41598-018-24400-4. Sci Rep. 2018. PMID: 29670134 Free PMC article.
References
- Collins FS, Finnell RH, Rossant J, Wurst W. A new partner for the international knockout mouse consortium. Cell. 2007;129:235. - PubMed
- Collins FS, Rossant J, Wurst W Consortium, T. I. M. K. A mouse for all reasons. Cell. 2007;128:9–13. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous