Synthetic spike-in standards for RNA-seq experiments - PubMed (original) (raw)
Synthetic spike-in standards for RNA-seq experiments
Lichun Jiang et al. Genome Res. 2011 Sep.
Abstract
High-throughput sequencing of cDNA (RNA-seq) is a widely deployed transcriptome profiling and annotation technique, but questions about the performance of different protocols and platforms remain. We used a newly developed pool of 96 synthetic RNAs with various lengths, and GC content covering a 2(20) concentration range as spike-in controls to measure sensitivity, accuracy, and biases in RNA-seq experiments as well as to derive standard curves for quantifying the abundance of transcripts. We observed linearity between read density and RNA input over the entire detection range and excellent agreement between replicates, but we observed significantly larger imprecision than expected under pure Poisson sampling errors. We use the control RNAs to directly measure reproducible protocol-dependent biases due to GC content and transcript length as well as stereotypic heterogeneity in coverage across transcripts correlated with position relative to RNA termini and priming sequence bias. These effects lead to biased quantification for short transcripts and individual exons, which is a serious problem for measurements of isoform abundances, but that can partially be corrected using appropriate models of bias. By using the control RNAs, we derive limits for the discovery and detection of rare transcripts in RNA-seq experiments. By using data collected as part of the model organism and human Encyclopedia of DNA Elements projects (ENCODE and modENCODE), we demonstrate that external RNA controls are a useful resource for evaluating sensitivity and accuracy of RNA-seq experiments for transcriptome discovery and quantification. These quality metrics facilitate comparable analysis across different samples, protocols, and platforms.
Figures
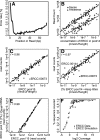
Figure 1.
Library characteristics, ERCC quantification, and coverage. Quality control plots for a stranded ENCODE RNA-seq library of K562 cell Poly-A+ RNA with ERCC spike-ins (library 7). (A) Mismatch rate along reads mapped to all ERRC RNAs. The first 6 bp correspond to the random reverse transcription hexamer-priming site. (B) Scatter plot for sense and antisense read counts per ERCC. (C,D) Scatter plots of read counts versus mass (concentration times length) per ERCC: (C) 100% ERCC library (library 6) and (D) pool of 44 2% ERCC spike-in H. sapiens libraries (libraries 7–50). ERCC-00073 showed aberrant abundance patterns in multiple RNA-seq experiments, as did ERCC-00144 in ERCC pool 14. They may have been inaccurately quantified in our ERCC test set due to errors during the complex mixing scheme used to generate the pools, as they are also suspect in RT-PCR and array experiments on these ERCC pools (M Salit, unpubl.). (E) Scatter plot of read counts in the 100% ERCC library versus a 1% ERCC spike-in D. melanogaster library (library 5). (F) Average sequencing depth and percentage of primary sequence covered for all ERCC transcripts for real data (black) and simulated data (gray).
Figure 2.
Estimation of cellular transcript abundance in a S2 cell. (A) This plot shows results from a library (library 3) made of 100 ng S2 polyA+ RNA (mRNA yield for this extraction is 0.175 pg/cell) and 5 ng of pool 15 ERCC RNAs. A linear regression of abundance estimated from RNA-seq and the known input amounts. Dashed lines represent 95% confidence intervals for the regression fit. (B) Distribution of S2 transcript abundance estimated from RNA-seq.
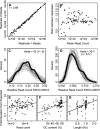
Figure 3.
Quantification errors and biases. (A) Scatter plot of read counts for each ERCC transcript in two different libraries of human RNA-seq with 2% ERCC spike-ins (K562 A+ Repl.1 and K562 A+ Repl.2, libraries 7, 8). (B) Scatter plot of fold deviation between replicates versus read counts for a given ERCC RNA. (C,D) Read counts for two example ERCCs relative to the total number of ERCC reads across 44 different libraries (libraries 7–50) with ERCC spike-in H. sapiens RNA samples (black line), the negative binomial distribution (solid gray), and random samples (n = 44) from the negative binomial distribution (dotted gray). The observed distribution fits a negative binomial model over a Poisson model (P < 2.2 × 10−16, likelihood ratio). (E_–_G) Scatter plots of the fold deviation between observed and expected read count for each ERCC in the 100% ERCC library (library 6) compared with read count (E), GC content (F), and ERCC RNA length (G).
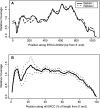
Figure 4.
Stereotypic read density heterogeneity in ERCC RNA-seq. (A) Traces of relative coverage along ERCC-0002 in two different ENCODE libraries (libraries 7, 8). The pattern is highly reproducible (Pearson's r = 0.96). (B) Average relative coverage along all control RNAs for ERCC spiked in the H. sapiens libraries (libraries 7–50). Dashed lines represent 1 SD around the average across different libraries.
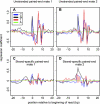
Figure 5.
Sequence patterns predictive of overrepresentation in RNA-seq. Patterns in the single-end 100% ERCC library (library 6) and ENCODE strand-specific pair end libraries (libraries 7–50) based on coefficients from the glm model (Li et al. 2010) (see Methods). (A,B) Regression coefficient for each base at positions around the beginning of reads mapped to the forward (A) and reverse (B) strands of ERCC-transcripts in the unstranded 100% ERCC library (library 6). (C,D) Regression coefficient for each type of nucleotide at different relative position to the upstream (C) or downstream (D) read of read pairs mapped to ERCC in the stranded ENCODE libraries. Adenosine is treated as base level in the regression model; i.e., the coefficient for “A” is always 0, while the other coefficients represent the predicted overrepresentation due to the presence of this nucleotide at this position, relative to an adenosine.
Figure 6.
Smoothing read densities. (A,B) Local read heterogeneity of a single ERCC RNA in the 100% ERCC library (library 6). Smoothing read density using the GLM linear model (B), and the more complex MART model (C; see text). (D) Variance in read depth of randomly drawn 50-bp windows from all ERCC RNAs based on an unbiased simulation, raw data, and the smoothed coverage from the sequence specific models. (E) The effect of coverage on read depth variance in simulated data. For the most abundant quartile of transcripts (Q1, mean coverage >19.9), the ratio of the read depth of 50-bp windows to the average depth is between 0.2853 and 1.2360. For Q2, (mean coverage >1.5), inner quartile range for the ratio is between 0.1917 and 1.2540. For Q3, (mean coverage >0.09), it is 0 with moderate large outliers (>21). For Q4 (mean coverage <0.08), it is 0 with very large outliers (>400).
Similar articles
- Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design.
Pine PS, Munro SA, Parsons JR, McDaniel J, Lucas AB, Lozach J, Myers TG, Su Q, Jacobs-Helber SM, Salit M. Pine PS, et al. BMC Biotechnol. 2016 Jun 24;16(1):54. doi: 10.1186/s12896-016-0281-x. BMC Biotechnol. 2016. PMID: 27342544 Free PMC article. - Normalization of human RNA-seq experiments using chimpanzee RNA as a spike-in standard.
Yu H, Hahn Y, Park SR, Chung SK, Jeong S, Yang I. Yu H, et al. Sci Rep. 2016 Aug 24;6:31923. doi: 10.1038/srep31923. Sci Rep. 2016. PMID: 27554056 Free PMC article. - Using Synthetic Mouse Spike-In Transcripts to Evaluate RNA-Seq Analysis Tools.
Leshkowitz D, Feldmesser E, Friedlander G, Jona G, Ainbinder E, Parmet Y, Horn-Saban S. Leshkowitz D, et al. PLoS One. 2016 Apr 21;11(4):e0153782. doi: 10.1371/journal.pone.0153782. eCollection 2016. PLoS One. 2016. PMID: 27100792 Free PMC article. - Statistical detection of differentially expressed genes based on RNA-seq: from biological to phylogenetic replicates.
Gu X. Gu X. Brief Bioinform. 2016 Mar;17(2):243-8. doi: 10.1093/bib/bbv035. Epub 2015 Jun 24. Brief Bioinform. 2016. PMID: 26108230 Review. - Current and Future Methods for mRNA Analysis: A Drive Toward Single Molecule Sequencing.
Bayega A, Fahiminiya S, Oikonomopoulos S, Ragoussis J. Bayega A, et al. Methods Mol Biol. 2018;1783:209-241. doi: 10.1007/978-1-4939-7834-2_11. Methods Mol Biol. 2018. PMID: 29767365 Review.
Cited by
- Revisiting global gene expression analysis.
Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. Lovén J, et al. Cell. 2012 Oct 26;151(3):476-82. doi: 10.1016/j.cell.2012.10.012. Cell. 2012. PMID: 23101621 Free PMC article. Review. - Erratum for Crossette et al., "Metagenomic Quantification of Genes with Internal Standards".
Crossette E, Gumm J, Langenfeld K, Raskin L, Duhaime M, Wigginton K. Crossette E, et al. mBio. 2021 Jun 29;12(3):e0117421. doi: 10.1128/mBio.01174-21. Epub 2021 Jun 1. mBio. 2021. PMID: 34061600 Free PMC article. No abstract available. - High-throughput full-length single-cell RNA-seq automation.
Mamanova L, Miao Z, Jinat A, Ellis P, Shirley L, Teichmann SA. Mamanova L, et al. Nat Protoc. 2021 Jun;16(6):2886-2915. doi: 10.1038/s41596-021-00523-3. Epub 2021 May 14. Nat Protoc. 2021. PMID: 33990801 - Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq.
Treutlein B, Lee QY, Camp JG, Mall M, Koh W, Shariati SA, Sim S, Neff NF, Skotheim JM, Wernig M, Quake SR. Treutlein B, et al. Nature. 2016 Jun 16;534(7607):391-5. doi: 10.1038/nature18323. Epub 2016 Jun 8. Nature. 2016. PMID: 27281220 Free PMC article. - Mediation of Drosophila autosomal dosage effects and compensation by network interactions.
Malone JH, Cho DY, Mattiuzzo NR, Artieri CG, Jiang L, Dale RK, Smith HE, McDaniel J, Munro S, Salit M, Andrews J, Przytycka TM, Oliver B. Malone JH, et al. Genome Biol. 2012 Apr 24;13(4):r28. doi: 10.1186/gb-2012-13-4-r28. Genome Biol. 2012. PMID: 22531030 Free PMC article.
References
- Baker SC, Bauer SR, Beyer RP, Brenton JD, Bromley B, Burrill J, Causton H, Conley MP, Elespuru R, Fero M, et al. 2005. The External RNA Controls Consortium: a progress report. Nat Methods 2: 731–734 - PubMed
- Bogdanova EA, Shagin DA, Lukyanov SA 2008. Normalization of full-length enriched cDNA. Mol Biosyst 4: 205–212 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous